You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000130_00091
You are here: Home > Sequence: MGYG000000130_00091
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
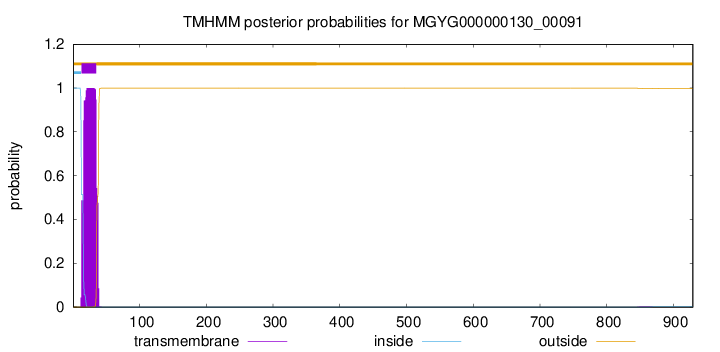
TMHMM annotations
Basic Information help
| Species | Peptoniphilus_C sp902363535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Peptoniphilaceae; Peptoniphilus_C; Peptoniphilus_C sp902363535 | |||||||||||
| CAZyme ID | MGYG000000130_00091 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83122; End: 85911 Strand: + | |||||||||||
Full Sequence Download help
| MGIKHTLRRK KTWQMLLAAI LFLLVLLGGI VAIWIVSILK DLPPVDFDDL TTSIPQTSYI | 60 |
| LDAEGNPIDE VDPSVFSENI SVDQVPDHVK EAFLAVEDRS FYKHHGLDFR QLGASILTNA | 120 |
| KSGAIERGGS TITQQLVKNV YLSSEQSLDR KIKEAYLTLG MEEKLGKDAI LEAYLNRVDL | 180 |
| GLGSQGIEAA SNAYFSKHAK DLTVEEGALL AGIVKSPASY QPIKRIPTEE NDGANVIATQ | 240 |
| TIGDRSYDLI ENPRAEERKK VVLRAMNQAG YITAEEAKSY AQIPISFMPK ENPPSPYSSY | 300 |
| VADVISDEAV AILAKISHMD KQAAEKQVRE GGLTIHSTIV GDFQKKMDAL YDTYPALIAR | 360 |
| GKNKGANFVD FNTDGNDRMI DEEGHVLYYP YEALFDEKGN MHLPAGAFTK TDGGLEIDGE | 420 |
| YFTESNGNLL LKNLYYLDEN GNLRTVAGAG TLFQAGDLKN PSTLLLQGEL YEAYKDAIEI | 480 |
| TEEELVLPAD LFLLPEKGSL QPQSAAIIAD NTTGAIVALA GGNDLKDPAR LRYNHLTSKR | 540 |
| QPGTAIVPLT TYLAALSEGD TLATSYDDTP MHVDGIIWPN ADSFYGYDVL ADGAANTRLA | 600 |
| ISGKILERYG FDPVLKNLNR LGLYKGERQD DAVKTPKENA KRHDMTYDAM AAGNFVDGMN | 660 |
| LMALTNSYAR IASPSVSDNY TVQKITDRKG RVLYEHGKTA AKDIHREDAL LRYALGRSPL | 720 |
| AENLRASGYD AFAVSGTNKY RADYFALGAT PRYTYGLWMG NDLQKIALAG DRSLTESLYA | 780 |
| SLVAILGDGT PWELPADFEM HEVSDKTGLL ATTYARRARS TVTLPFLPNT APTKETQNYT | 840 |
| RKLICSVSGQ LASTYCPYET ITYGYYFVRP KGYDPKAFDG ILPKDYYTLP SNYCQVHTKE | 900 |
| WYDQETQKAN EDEQDQDNDN DQNRDTHRR | 929 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 66 | 266 | 7.3e-58 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 1.15e-92 | 9 | 784 | 9 | 586 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 1.58e-90 | 9 | 835 | 3 | 767 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 2.20e-88 | 78 | 762 | 3 | 502 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| pfam00912 | Transgly | 7.87e-65 | 64 | 267 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| TIGR02071 | PBP_1b | 4.72e-45 | 80 | 762 | 150 | 658 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEJ35778.1 | 0.0 | 2 | 912 | 3 | 911 |
| VEJ34688.1 | 1.98e-148 | 17 | 904 | 24 | 924 |
| QQE47034.1 | 1.98e-148 | 17 | 904 | 24 | 924 |
| QQT90658.1 | 6.14e-147 | 17 | 904 | 24 | 924 |
| QQY80508.1 | 1.06e-130 | 27 | 900 | 40 | 937 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 2.11e-102 | 39 | 890 | 2 | 865 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 3NB6_A | 6.90e-42 | 80 | 276 | 23 | 193 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 9.41e-42 | 80 | 276 | 23 | 193 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3DWK_A | 8.79e-34 | 55 | 773 | 5 | 567 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 4OON_A | 2.27e-33 | 85 | 695 | 47 | 574 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O66874 | 1.51e-47 | 33 | 811 | 21 | 695 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| A5I6G4 | 1.63e-45 | 1 | 857 | 1 | 720 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7FY32 | 1.63e-45 | 1 | 857 | 1 | 720 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 5.07e-45 | 1 | 857 | 1 | 720 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| Q891X1 | 2.25e-42 | 37 | 762 | 41 | 629 | Penicillin-binding protein 1A OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
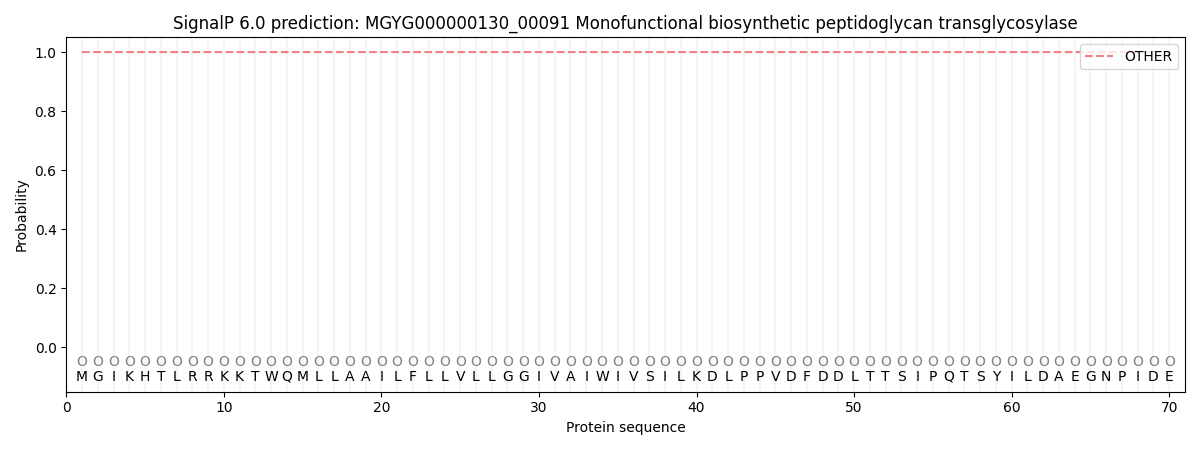
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000039 | 0.000005 | 0.000000 | 0.000000 | 0.000000 | 0.000001 |