You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000806_00011
You are here: Home > Sequence: MGYG000000806_00011
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
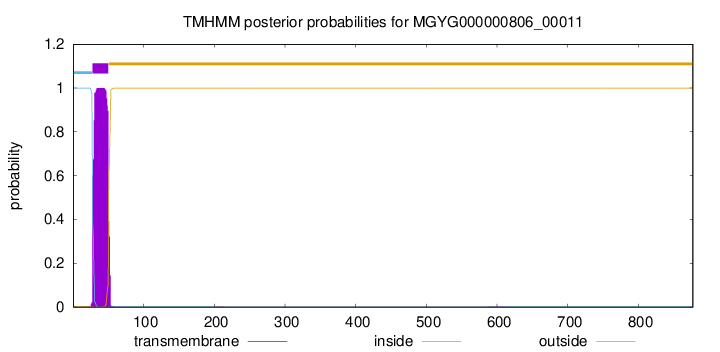
TMHMM annotations
Basic Information help
| Species | Blautia_A sp900541345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp900541345 | |||||||||||
| CAZyme ID | MGYG000000806_00011 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10571; End: 13204 Strand: + | |||||||||||
Full Sequence Download help
| MNYGKKATSK KRNSLISRTS MMGKRAHVSF IRVLFISLIA ICIMGACLGI GSFKGVIDNA | 60 |
| PDVDDIDIMP LGYASFLYDD HGNQLRKLAA PDSNRLPVSI EQIPEDLQHA VVAIEDERFY | 120 |
| EHNGIDVRGI LRAGVKAITS GNISEGASTI TQQLLKNNVF TTWTSESTWI EKFTRKFQEQ | 180 |
| YLAIQVEEKI GDKNVILENY LNTINLGAGA YGVQAAARQY FNKDVWDLNL SECATLAGIT | 240 |
| QNPSKFNPIE NPKDNAKRRK EVLQHMLEQG YIDQAKYEEA LNDDVYSRIQ AAQAETSSSR | 300 |
| NKVYTYFEDE LTDQIINDLM NIKGYTKAQA RNLLYSGGLK IMTTQDPNIQ KILDEEYSNP | 360 |
| ENYPANTQYA LDYALTVTNP EGNEVNYSKE MLKLFFQNED PEFDLLFASQ EEGQTYVDRY | 420 |
| KASILADGSK VVAERVNFAP QPQSSMSVID QHTGYVKAII GGRGEKTASL TLNRATDTTR | 480 |
| QPGSTFKIVS TYAPALNEMG MSLATTYEDK EIFYPDGSPV NNYSRTYSDS WMTIRQAIRS | 540 |
| SINTVAVQCL EDVTPELGLK YLDNFGFTTL AHGEADENGN IYTDANLSTA LGGITSGVTN | 600 |
| VELCAAYASI ANKGNYIKPI YYTQILDHNG NVLIDNTSAE RSVIKESTAW LLTNAMEDVV | 660 |
| QNGTGRACQL DNMPVAGKTG TTDDYNDLWF VGYTPYYTCA VWSGFDNNEK LPEDARNFHK | 720 |
| NLWRKVMSRI HEGLEYKEFE QPASVENTSI CEETGLLPRA GCPVITEYFD VGTLPTEYCD | 780 |
| QHFYEPPEEE FIEYEVTPTP EGEMPNPENP DGNGENQGGG DGSGENPGGD DGSGEYPGGD | 840 |
| DGSGENQGGG DGSGENSGGD DGSGENSGGD DDIIYYD | 877 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 85 | 266 | 3.1e-62 | 0.9661016949152542 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 7.70e-176 | 94 | 733 | 1 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 1.01e-152 | 30 | 742 | 10 | 608 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 8.80e-149 | 31 | 821 | 4 | 795 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 4.00e-77 | 81 | 267 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG4953 | PbpC | 1.15e-75 | 31 | 788 | 5 | 615 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL24654.1 | 0.0 | 1 | 785 | 1 | 787 |
| QCU02608.1 | 0.0 | 1 | 784 | 1 | 785 |
| QBE99737.1 | 0.0 | 1 | 790 | 1 | 792 |
| QIB57705.1 | 0.0 | 1 | 790 | 1 | 792 |
| QMW77137.1 | 0.0 | 1 | 790 | 1 | 792 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 1.31e-98 | 59 | 778 | 9 | 810 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 4OON_A | 1.04e-67 | 77 | 763 | 23 | 755 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3DWK_A | 3.16e-57 | 77 | 748 | 10 | 605 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 3ZG8_B | 1.87e-56 | 188 | 754 | 4 | 478 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3UDF_A | 6.30e-55 | 96 | 706 | 40 | 689 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0SRL7 | 1.05e-108 | 30 | 782 | 32 | 724 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain SM101 / Type A) OX=289380 GN=pbpA PE=3 SV=1 |
| Q0TNZ8 | 2.59e-108 | 30 | 782 | 32 | 724 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 6.90e-106 | 24 | 782 | 7 | 702 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 1.03e-105 | 24 | 782 | 7 | 702 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7FY32 | 1.03e-105 | 24 | 782 | 7 | 702 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
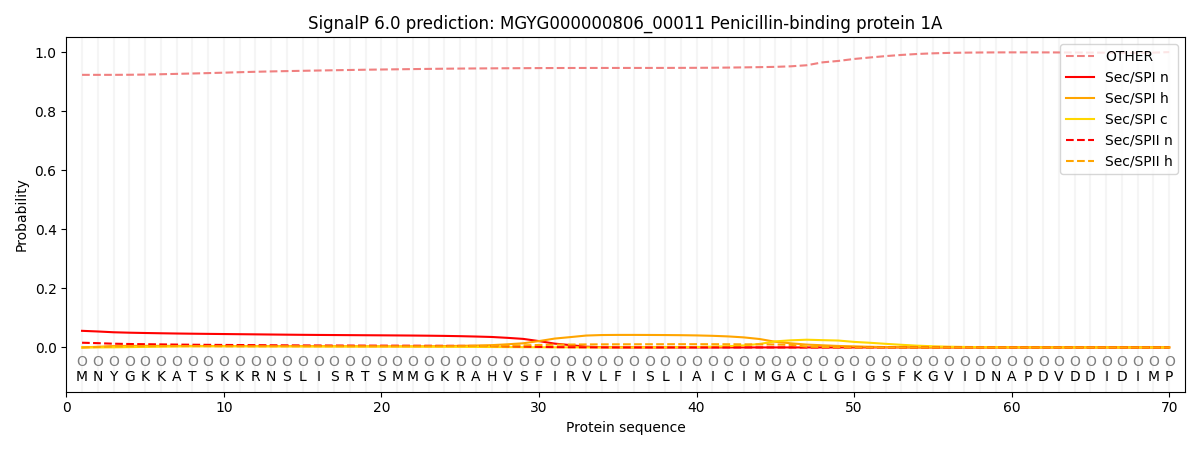
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.925344 | 0.049996 | 0.018348 | 0.000391 | 0.000261 | 0.005651 |