You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001159_00012
You are here: Home > Sequence: MGYG000001159_00012
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
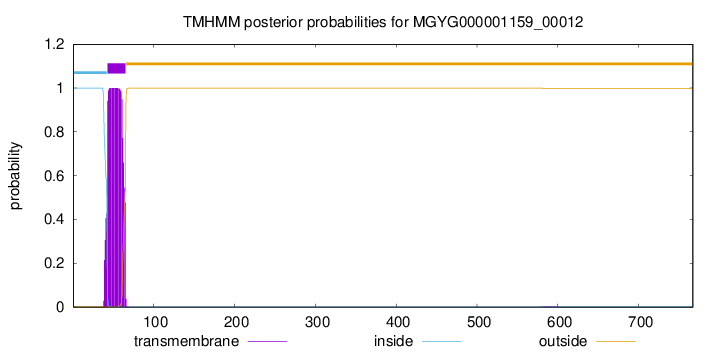
TMHMM annotations
Basic Information help
| Species | Bifidobacterium pullorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium pullorum | |||||||||||
| CAZyme ID | MGYG000001159_00012 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 131; End: 2434 Strand: - | |||||||||||
Full Sequence Download help
| MVSQTRTAKR TPAARHFGGG IHRSHGSKGP KPKGKSKHLI LKWVLGIIGA LLALGIGAFA | 60 |
| YLYATTEIPQ PESIAVAENT TVYYADGTTP IGTFSEQNRE IIDCSVLPDY VGQAVVASED | 120 |
| RSFYTNRGID LVGIARAFWN NLTTGSRQGG STITQQYAER YYLGETTSYL GKAREAILAL | 180 |
| KIAQAQDKDQ VLCNYMNTIY LGRGTYGIQA AAKAYFGKDA KDLTMPEAAM IAGIIPSPSS | 240 |
| WDPAVDPEQA QARFTRVLRI MQEDGYITAQ EQQEVQFPQT IEYTQQNSYQ GANGYLLQMV | 300 |
| RDELTGDGTF SAEQLDTGGY AIVTTIDKSK QDLMYSVVSP AQNGMQGVIP DGMEFGGISV | 360 |
| NAKDGSIISV YAGEDYLTKQ LNQATQSVYE IGSTMKPMAL LGAIQEGVNL DTVFNGNSPR | 420 |
| KFDGIADPVG NFGNMSYGNV NLYTATAQSL NTVYMDVQAK LGTQRIAEIA KEAGAESDAL | 480 |
| DGTNPFTVLG NNALTTKDVA RMYVTIANQG NRPNIHIVSS VKNTDGEDIY KAPTETTQVF | 540 |
| DANDTALVTK AMTGTVQNGT ATEALAVGHN LAMKTGTAND SYAASAVGFT PSVVSVFAMW | 600 |
| YPDANGNPQE VPAFGGWSGG SDYPVHLFTQ YMTQALAGTD NETFPNAVDH GKVGGPDGSW | 660 |
| GLGSQTLQQM QGAQRKAEEE AQRQAEEEAQ RQAEEEAQRQ AEEEARRQQE QAEQERLEYC | 720 |
| QNNPNDVSCG GSGNTSGNTS GNEGAGEESG DGSEVEGGET DDDSEQS | 767 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 89 | 262 | 6.4e-61 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 8.28e-131 | 45 | 645 | 18 | 606 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 1.28e-127 | 98 | 638 | 1 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 4.81e-108 | 40 | 644 | 4 | 727 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 4.70e-78 | 89 | 262 | 3 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG4953 | PbpC | 2.02e-47 | 46 | 616 | 2 | 546 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAY33663.1 | 0.0 | 1 | 766 | 1 | 758 |
| QTL79949.1 | 0.0 | 22 | 679 | 35 | 692 |
| QTL78070.1 | 0.0 | 22 | 679 | 35 | 692 |
| VEG22900.1 | 0.0 | 22 | 679 | 35 | 692 |
| ADB08925.1 | 0.0 | 22 | 679 | 35 | 692 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5U2G_A | 6.91e-40 | 64 | 532 | 3 | 566 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 5FGZ_A | 4.59e-39 | 89 | 633 | 146 | 696 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 5.14e-39 | 89 | 633 | 167 | 717 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3DWK_A | 7.80e-39 | 97 | 648 | 25 | 602 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 3NB6_A | 2.06e-38 | 92 | 274 | 13 | 196 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P71707 | 2.37e-74 | 59 | 655 | 152 | 765 | Penicillin-binding protein 1A OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ponA1 PE=1 SV=3 |
| A0R7G2 | 6.08e-72 | 34 | 655 | 61 | 703 | Penicillin-binding protein 1A OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=ponA1 PE=3 SV=1 |
| O66874 | 1.29e-59 | 42 | 644 | 3 | 676 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P0A0Z5 | 6.29e-59 | 40 | 638 | 5 | 708 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=mrcA PE=3 SV=1 |
| P0A0Z6 | 6.29e-59 | 40 | 638 | 5 | 708 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup B (strain MC58) OX=122586 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
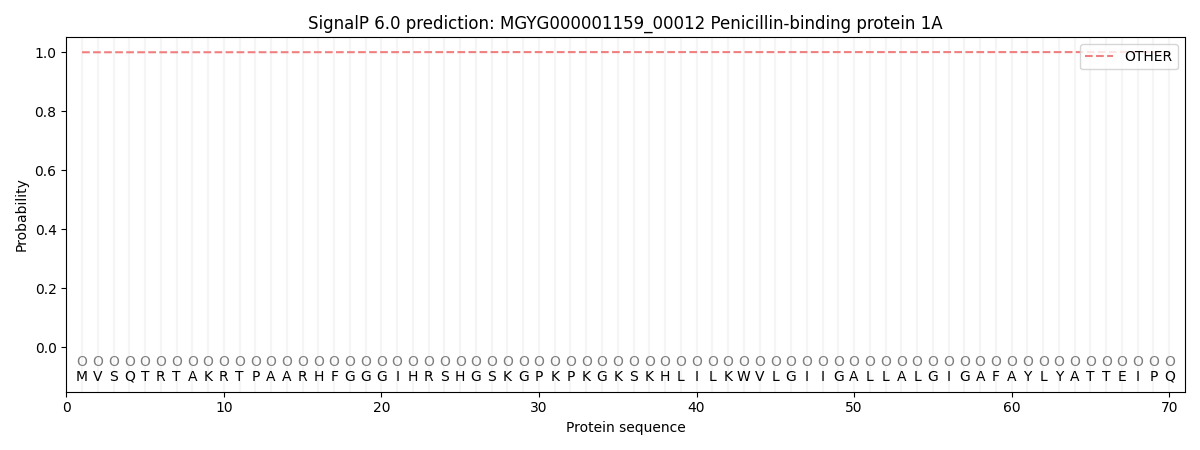
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999730 | 0.000279 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |