You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001359_00411
You are here: Home > Sequence: MGYG000001359_00411
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
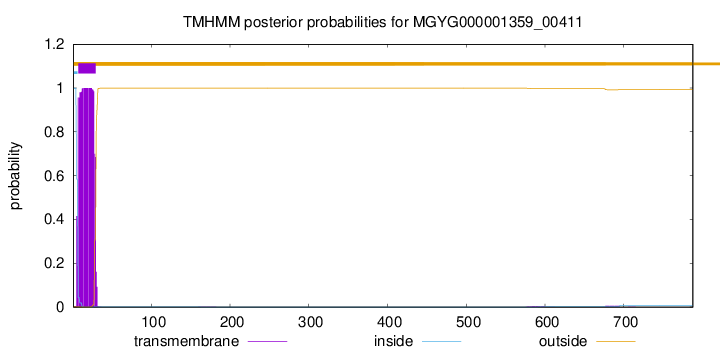
TMHMM annotations
Basic Information help
| Species | Bilophila wadsworthia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Bilophila; Bilophila wadsworthia | |||||||||||
| CAZyme ID | MGYG000001359_00411 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 436259; End: 438625 Strand: - | |||||||||||
Full Sequence Download help
| MKRVFFLFGT IIGVLVALGL AAVVGVYFWA VRDLPGFTRI ADYRPALTTT VLARDGSVIG | 60 |
| SFYRENRFLI SLDQMPKMLP KAFLAAEDAE FYEHEGVNPV AIFRAFLINL QSGTKRQGGS | 120 |
| TITQQVIKRL LLTPERSYER KIKEAILAYR LEHYLSKDEI LTIYLNQTFL GANAYGVEAA | 180 |
| ARTYFGKNAK DLTLAECALI AGLPKAPSQY NPYRDPSAAI GRQHYVLRRL RELNWITEAE | 240 |
| YDAALKQPLV FKQMADGMGR QGAWYLEEVR RQLIDLFSEE NAKRLGIELP VYGEDAVYQM | 300 |
| GFTVQTAMSP PAQMAADEAL RSGLEDFTRR QGWQGPVEKI PVADLKERIE KSHFSPDMLA | 360 |
| NGERVKGIVT RITDKGADIR LGKYSGFIDN AQLSWAKKTL RSKQKFLESG DVVWVAAVSD | 420 |
| PKNPYNPDEA ELNKPIRLSL QCVPELQGAL ISIEPETGDV VAMVGGYSFQ ESQFNRATQA | 480 |
| FRQPGSSFKP IVYSAALDNG FTAASVILDA PVVQFLESGD VWRPSNYEKN FKGPLLLRTA | 540 |
| LALSRNLCTI RVVQQMGVQK VIERAKDLEL EPHFPEVLSV SLGAVAVTPL NMTQAYTAFA | 600 |
| NGGMVSKARF ILSVKNFWNE TIYESQPDLR DAISPQNAYV MSYLLKEVVN AGTATKAKVL | 660 |
| GRPVAGKTGT SNDWKDAWFI GFTPHLVTGM YVGYDQPRTM GRSGTGGSMA LPIFVEYAKS | 720 |
| AFQAYPPDDF EVPDGISFAN VDQTSGHLVG SGGLRLPFYT GTEPGSSTSE EAISGVNTRG | 780 |
| EDLLKQMF | 788 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 55 | 230 | 4.3e-69 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 0.0 | 66 | 723 | 1 | 530 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 0.0 | 1 | 772 | 8 | 642 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 0.0 | 1 | 771 | 2 | 771 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 1.35e-154 | 1 | 779 | 1 | 839 | penicillin-binding protein 1a; Provisional |
| TIGR02071 | PBP_1b | 1.03e-115 | 65 | 764 | 145 | 726 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGC49761.1 | 0.0 | 4 | 788 | 9 | 804 |
| ABE16187.1 | 0.0 | 4 | 788 | 17 | 812 |
| AAR40712.1 | 0.0 | 4 | 788 | 17 | 812 |
| CAJ54403.1 | 0.0 | 4 | 788 | 9 | 804 |
| AAS95114.1 | 0.0 | 16 | 788 | 16 | 807 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OON_A | 5.66e-134 | 51 | 752 | 23 | 753 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3UDF_A | 1.47e-120 | 51 | 726 | 23 | 720 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 5U2G_A | 4.94e-109 | 30 | 764 | 3 | 800 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 7U4H_A | 3.49e-77 | 48 | 764 | 20 | 807 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 5FGZ_A | 1.04e-65 | 36 | 760 | 136 | 730 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q1RKC5 | 3.22e-165 | 2 | 758 | 3 | 753 | Penicillin-binding protein 1A OS=Rickettsia bellii (strain RML369-C) OX=336407 GN=mrcA PE=3 SV=1 |
| Q4UK08 | 4.27e-155 | 2 | 752 | 3 | 747 | Penicillin-binding protein 1A OS=Rickettsia felis (strain ATCC VR-1525 / URRWXCal2) OX=315456 GN=mrcA PE=3 SV=1 |
| O87626 | 8.33e-154 | 34 | 736 | 35 | 719 | Penicillin-binding protein 1A OS=Neisseria flavescens OX=484 GN=mrcA PE=3 SV=1 |
| Q68VU2 | 6.32e-153 | 1 | 752 | 1 | 747 | Penicillin-binding protein 1A OS=Rickettsia typhi (strain ATCC VR-144 / Wilmington) OX=257363 GN=mrcA PE=3 SV=1 |
| Q9ZCE9 | 6.69e-153 | 1 | 752 | 1 | 747 | Penicillin-binding protein 1A OS=Rickettsia prowazekii (strain Madrid E) OX=272947 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
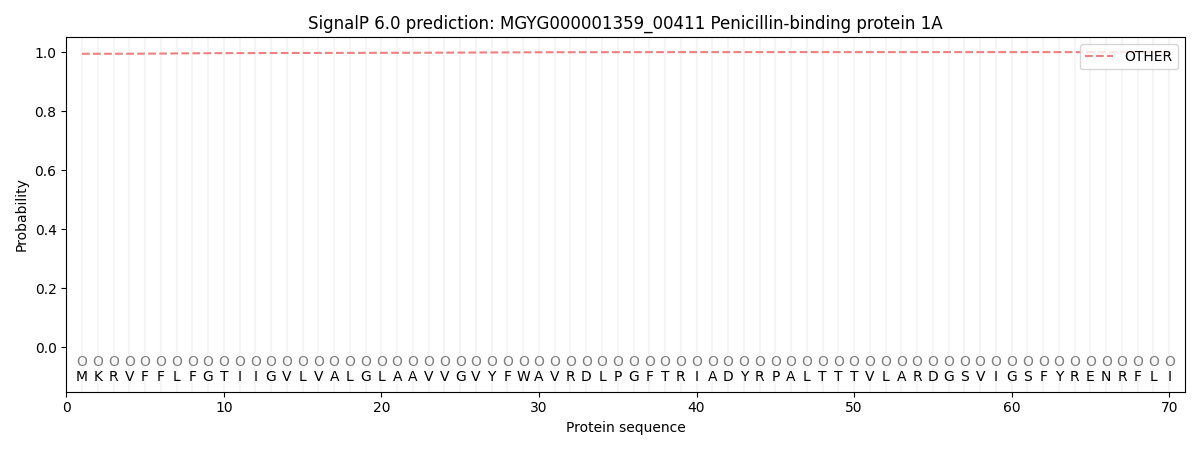
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.994502 | 0.002756 | 0.000923 | 0.000017 | 0.000010 | 0.001818 |