You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001953_01485
You are here: Home > Sequence: MGYG000001953_01485
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
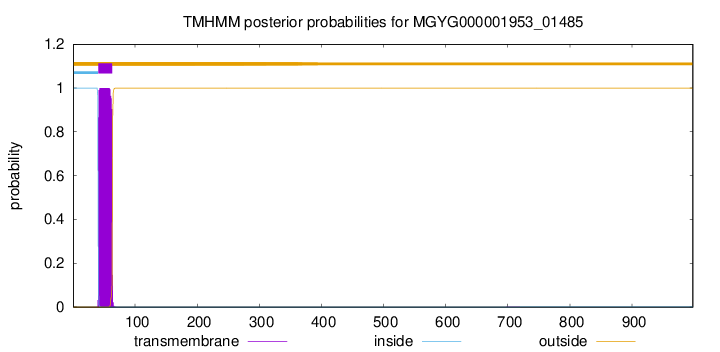
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; ; | |||||||||||
| CAZyme ID | MGYG000001953_01485 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1187; End: 4183 Strand: + | |||||||||||
Full Sequence Download help
| MRSDKRKEKK KAKKHASTET AVVNSDGTAS RKKYRLNKKR FFLFLLTLIL IAIIGVAIYV | 60 |
| GVIISQAPKI DTDDIYTILT ESTVIYDDDG KEIDTVYTEA NRENVEYKEL PENLVNAFVA | 120 |
| LEDKTFWDHH GFNIVRIFGA IKDSFTSGHI SGTSTITQQL ARNLFLKEKM TEHTLERKII | 180 |
| EAYYAIILEK ELSKEEIMEA YLNTVYLGYQ SNGVQAASQA YFSKDVQDLS LAQCAALASL | 240 |
| VQAPSDYQLV ELVGNDDVST KDTNIIKRTS AGTYIANDAS KNRRETCLKL MKEQGYISQE | 300 |
| EYDKAVSVSL KDMLNPSYNS SSTEASYFED YVIEEVINDL MEANDWDYNT AWEKVYNGGV | 360 |
| KIYSTLDSQA QNVITTEFEN DTNFPGVSIS TDGNGNIVNK YGQISLYAYS NYFDDDGNFT | 420 |
| FTNDEITRQD DGSLIIKAGK RLNIYDTEVN GETDYSIEFK NLYTMENGTL YAISGGYINI | 480 |
| PQQYKSKNKN GNIVVSAEFF EDEQYKDFFI FNDDGTVTVP SRSYTLNQKV IQPQAAMTIV | 540 |
| DNATGQIKAM VGGRKTTGRQ LYNRATNPRQ PGSSIKPLSV YAAAIQQSAD EAANGQKHKF | 600 |
| VDYNIDEQGA DLYGNYLTAG SIVIDEETTI NGRVWPRNSG GGYSGPQTMR SALQLSINTC | 660 |
| AVKILQQVGT DYSADLVEKF GITTLDREGS TNDINLAALG LGGMSSGVTT LEMASAYTVF | 720 |
| PNNGTRQETT SYTKVVDTHG NTLLDNSQTE THEVLNSGVA WIMADMLKSV VTSGLGSPAA | 780 |
| ISGVQVGGKT GTTDDEYDIW FDGFTPSYSA SLWIGNDQNF TLTSLSSYAA RLWGRIMNQI | 840 |
| DGAKTGSYKD MPSNVISSGG EYYISGTQGG TGSLDDLEKE VTICTDSGYL ATPSCTHTET | 900 |
| VTYSTYGDEA EEIPEYYCYM HNPDTEKYPI SPDETLQPQD PIKPPDEEDP ETPPDEGGDG | 960 |
| GGDNPNGGDE GGDSGQGGGN QGSGDGGNQG GGNTPSTQ | 998 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 89 | 254 | 1.3e-56 | 0.8983050847457628 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 1.04e-154 | 101 | 840 | 1 | 528 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 4.32e-146 | 82 | 871 | 55 | 625 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 9.92e-128 | 55 | 868 | 22 | 763 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 9.38e-71 | 85 | 868 | 50 | 827 | penicillin-binding protein 1a; Provisional |
| pfam00912 | Transgly | 3.52e-66 | 89 | 247 | 1 | 156 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKT47092.1 | 0.0 | 15 | 931 | 4 | 922 |
| ASS40664.1 | 0.0 | 50 | 867 | 76 | 892 |
| ASS37973.1 | 1.71e-308 | 20 | 870 | 97 | 943 |
| AVM47767.1 | 3.45e-305 | 48 | 870 | 122 | 943 |
| QHI73342.1 | 8.58e-285 | 43 | 931 | 1 | 917 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 6.64e-128 | 65 | 856 | 3 | 776 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 3UDF_A | 4.82e-54 | 105 | 822 | 42 | 694 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 3ZG8_B | 7.80e-54 | 188 | 838 | 3 | 458 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 4OON_A | 1.52e-51 | 85 | 822 | 23 | 701 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3DWK_A | 6.62e-47 | 85 | 868 | 10 | 615 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q97GR5 | 1.93e-93 | 39 | 876 | 30 | 702 | Penicillin-binding protein 1A OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 3.89e-93 | 46 | 858 | 21 | 661 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A7FY32 | 1.10e-92 | 46 | 858 | 21 | 661 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 1.10e-92 | 46 | 858 | 21 | 661 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| Q8XJ01 | 8.46e-86 | 24 | 837 | 14 | 662 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
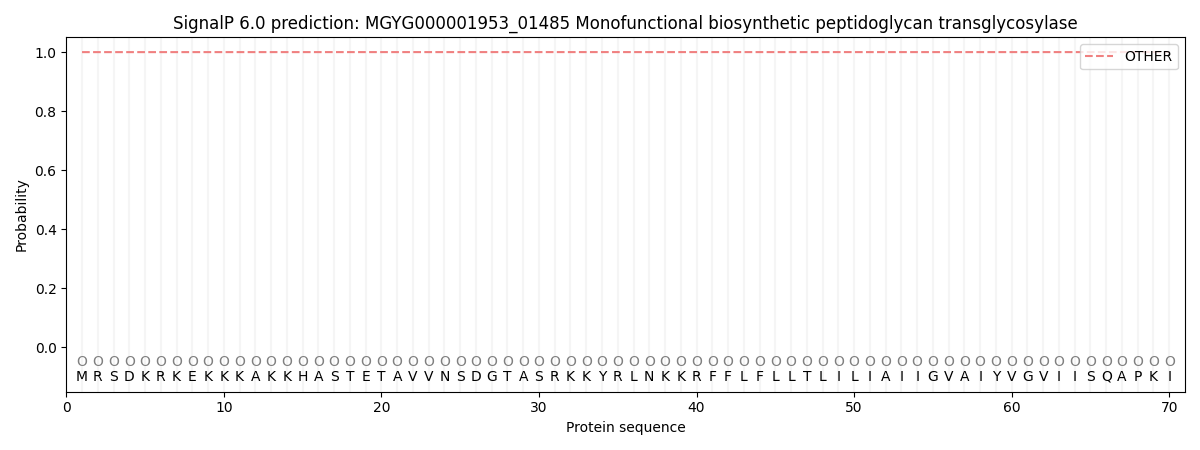
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999977 | 0.000032 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |