You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002619_00072
You are here: Home > Sequence: MGYG000002619_00072
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
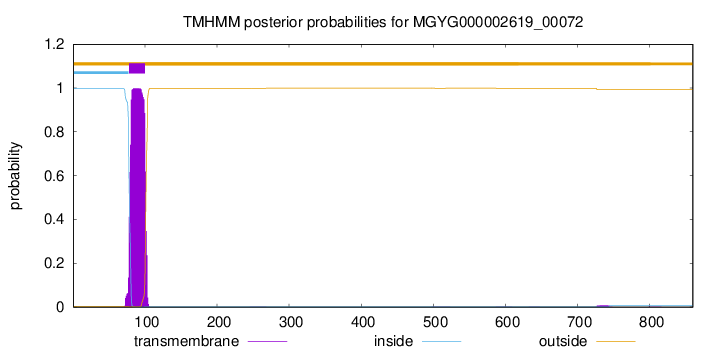
TMHMM annotations
Basic Information help
| Species | Faecalibacterium prausnitzii_J | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Faecalibacterium; Faecalibacterium prausnitzii_J | |||||||||||
| CAZyme ID | MGYG000002619_00072 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 89441; End: 92023 Strand: - | |||||||||||
Full Sequence Download help
| MKCQEAIPIS NNEMRKIHRG GEEQPARPMG RKVNVSQSRR EVRPAAENRT RAPKAPKTPK | 60 |
| PPKEKSKHPM LRFIGRTLAT VLCLGIMLGS VVAVGIVFYV VQATANDGDL LDLDNIQLSQ | 120 |
| SSMVVATDPD TGAQVEYATL RSSNSHRVWA DLEQIPSNLQ YAFICTEDKD FYSEPGVNFK | 180 |
| RTIGAMINEY LLPIYSSKQG ASTLEQQLIK NLTDDNSASG IDGALRKVRE IYRALCLSRS | 240 |
| YSKETILEAY LNTISFTGTI QGVQTAANEY FNKDVSQLSL WECATIASIT KNPTNYNPYT | 300 |
| NPENLINRRN FIMYNMWQQG VISEEDYRNG AAQPLVLAEE DSGKKPSTVT SYFTDALFSE | 360 |
| VVQDIMVKEG ISESEAQKLL YTGGFTIEAT VNTKLQSQME NLMLNTDDAY FPAGWHEEEV | 420 |
| TSISDDDVQV FNEDGTPKTR TGEDGTVYYY RNVRTQAAMV TLDYDGNVLA IVGGLGEKTK | 480 |
| SLSLNRAYNV ERQTGSTIKP IGAYALGIEY GLVNWSTMLN NSPLYQKQDM IIRDEDYCRK | 540 |
| NGLMGLSDKQ LRAYPNAWRS WPRNYGGNYG DGSDIPLWNG LARSLNTIAI RVGDLVGASN | 600 |
| IFNFVYNTLQ LNTLDPTNDV GLAQMVMGSQ TKGVTPMALA AAFQIFYDGE YTTPHLYTRV | 660 |
| LDRDGNIYLE NNATSYQALT PDTAYVMNRL LKNVLFSSVG TASGRYPNSN GMEAFGKTGT | 720 |
| ASDEKDLWFV GGTPYYVTAV WWGYDAPYDM TKTLSKQQAK TRTCVTAWKA LMEQAQADLP | 780 |
| YKAFPTSAGV VERRYCTQSG LLAGAGCPST AVGYYRADDL PDTCTYSHAA PQAAVSTENT | 840 |
| DGAVPTQPVV PTDTSALDTE | 860 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 143 | 316 | 4.7e-54 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 2.23e-129 | 72 | 785 | 9 | 606 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 4.14e-126 | 146 | 775 | 1 | 528 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 2.74e-104 | 70 | 815 | 1 | 758 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02071 | PBP_1b | 4.72e-64 | 138 | 745 | 137 | 658 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| pfam00912 | Transgly | 1.07e-58 | 146 | 316 | 12 | 176 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXB28952.1 | 0.0 | 14 | 860 | 1 | 847 |
| CBL02306.1 | 0.0 | 29 | 860 | 1 | 832 |
| QIA44319.1 | 0.0 | 17 | 860 | 1 | 833 |
| ATL89188.1 | 0.0 | 33 | 860 | 1 | 816 |
| ATO98888.1 | 0.0 | 33 | 860 | 1 | 816 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 4.71e-53 | 146 | 815 | 39 | 801 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 4OON_A | 1.23e-45 | 155 | 821 | 47 | 768 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3ZG8_B | 1.05e-41 | 237 | 745 | 3 | 433 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3UDF_A | 1.54e-41 | 151 | 780 | 43 | 720 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 3DWK_A | 2.05e-39 | 139 | 744 | 20 | 557 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q97GR5 | 7.84e-81 | 73 | 792 | 28 | 683 | Penicillin-binding protein 1A OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 3.36e-77 | 143 | 802 | 72 | 670 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 1.75e-76 | 143 | 802 | 72 | 670 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| A7FY32 | 1.75e-76 | 143 | 802 | 72 | 670 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| Q891X1 | 1.58e-75 | 64 | 826 | 10 | 712 | Penicillin-binding protein 1A OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
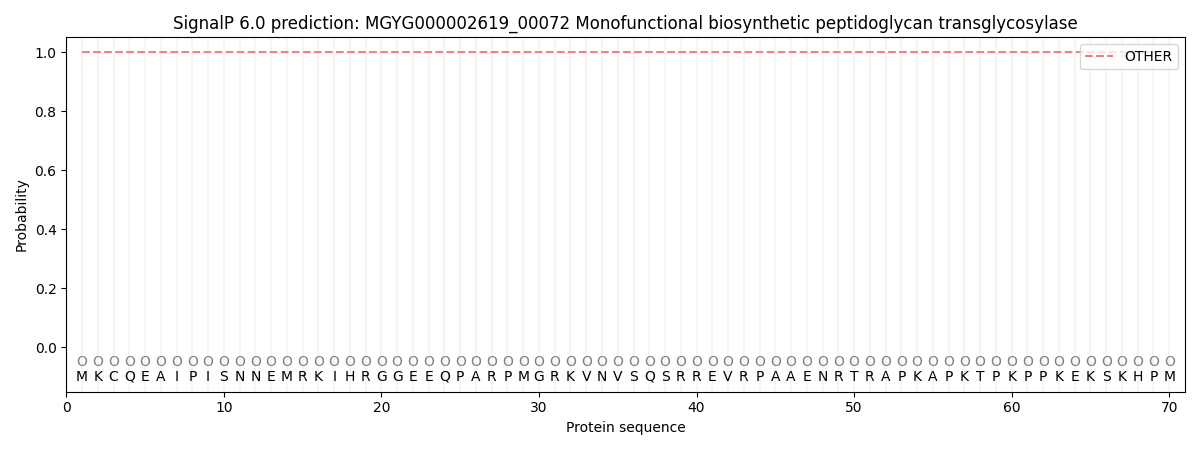
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000072 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |