You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002987_00073
You are here: Home > Sequence: MGYG000002987_00073
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
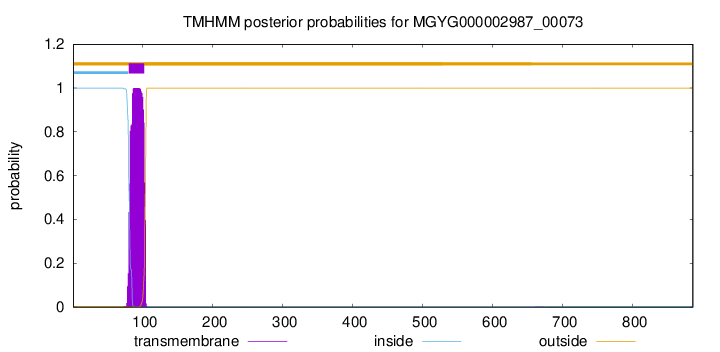
TMHMM annotations
Basic Information help
| Species | TM7x sp900555265 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Patescibacteria; Saccharimonadia; Saccharimonadales; Saccharimonadaceae; TM7x; TM7x sp900555265 | |||||||||||
| CAZyme ID | MGYG000002987_00073 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional glycosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61366; End: 64026 Strand: + | |||||||||||
Full Sequence Download help
| MKKKTTISTA GHGSSDKKSP SKRMNLYANL AQKHKTKKDK DARERAEYLA TLPKHPVKRF | 60 |
| FYRLHPKRLA KYWFSKRGGL MALKILGVSI LLMLLLIGGM FAYFRKDLDK IRPGELAKRV | 120 |
| QTTVTKYYDR NDNLLWEDKG TGNYQLVVES DQISDYLKKA TVAIEDRNFY NHHGISISGM | 180 |
| LRAMLSTASR RQVQGGSTLT QQLVKQVFFA DEAGDRSISG IPRKIKEIIL AIEVERMYSK | 240 |
| DQILSLYLNE SPYGGRRNGA ESAAQTYFGK HAKDLTLAEA ALIAGIPQNP TYYNPYNTAG | 300 |
| NKALIARQHT VLDYMAEQGV ITKDEAEKAK KIDILSTLKP QTEHLENIKA PHFLLMVRNQ | 360 |
| LSKELGESVV GRGGLTIKTT LDWRIQEKLE NEMKSFFATG RPDAVRISNG AATVEDVQTG | 420 |
| QIVALVGSRD FNYTGFGQDN AAVSYIQPGS TIKSLVFAQL FDKHDSKQAY GSGTVLADEN | 480 |
| IDKLYGATLR NWDNKFMGAI NIRRALALSR NVPAIKAAYI VGDGSAKPVV EGIRRMGDTN | 540 |
| YCRQEENAGG YGLGAAIGAC GTKQTELVNA YSTLARMGVQ KDISSVIEVK NSQGETLKKW | 600 |
| KDEGKQVIDS QSAYIVNDIL SDRTPGLHGW MGVNGVRTSA KTGTSDKGSQ PKDLWIINYS | 660 |
| PALVMGMWLG NSDTSVIGTS ASNYGMPVIR SVMEFAHTQV YAKEGKWKSG QWYERPSGIQ | 720 |
| TVNGELYPSW WNKRQSQSTE KITFDKVSKK KATNCTPDGA REEIEVTKII DPLTKKESIT | 780 |
| VPSGYDANAE DDVHKCDDTK PQIGAISYTN SGKKYTINVD VTAGTWGLSA IEITVDGKSI | 840 |
| KSSEITSSGK QTATVEFDTT GAHTVSVTVR DSAYYTATST GSIQVN | 886 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 144 | 315 | 4.5e-60 | 0.9265536723163842 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 4.56e-142 | 101 | 697 | 32 | 597 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 3.17e-133 | 148 | 697 | 5 | 529 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG4953 | PbpC | 7.12e-95 | 101 | 679 | 22 | 543 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 3.92e-93 | 151 | 723 | 73 | 737 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02071 | PBP_1b | 8.54e-73 | 145 | 699 | 147 | 685 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT41586.1 | 0.0 | 1 | 884 | 1 | 884 |
| AJA06839.1 | 0.0 | 1 | 884 | 1 | 884 |
| QLF51933.1 | 0.0 | 20 | 885 | 25 | 895 |
| QJU11046.1 | 0.0 | 20 | 885 | 25 | 895 |
| QJU07589.1 | 0.0 | 20 | 885 | 25 | 895 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DWK_A | 9.01e-55 | 126 | 719 | 9 | 602 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 3.66e-51 | 117 | 719 | 9 | 611 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
| 2JE5_A | 8.04e-45 | 148 | 694 | 57 | 635 | StructuralAnd Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6],2JE5_B Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6] |
| 3ZG8_B | 3.80e-41 | 233 | 674 | 2 | 435 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 7U4H_A | 3.20e-40 | 129 | 670 | 25 | 734 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39793 | 5.21e-60 | 76 | 839 | 29 | 781 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| P70997 | 2.10e-58 | 117 | 696 | 46 | 607 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| A7GHV1 | 9.40e-58 | 133 | 723 | 57 | 662 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A7FY32 | 2.01e-56 | 133 | 723 | 57 | 662 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 2.01e-56 | 133 | 723 | 57 | 662 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
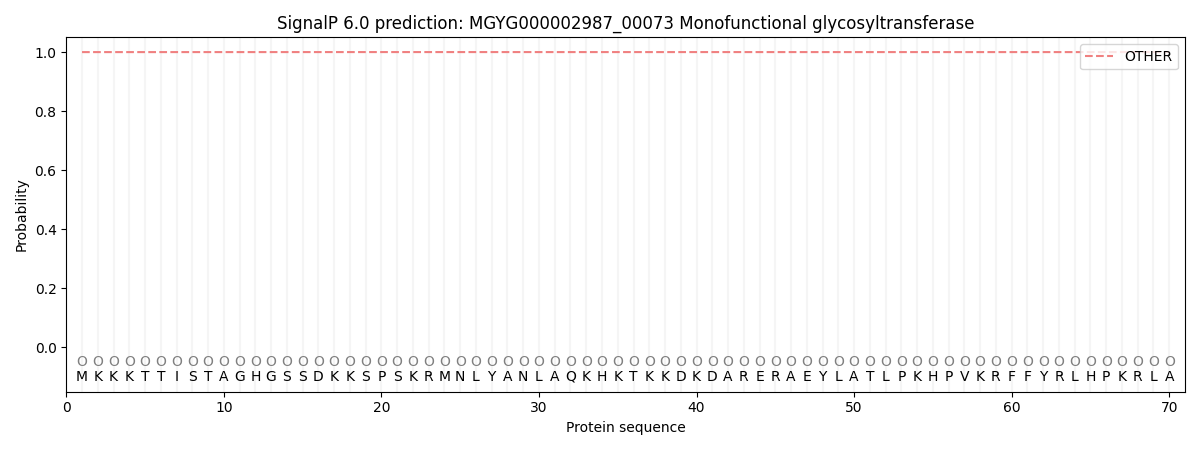
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000066 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |