You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003140_00243
You are here: Home > Sequence: MGYG000003140_00243
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
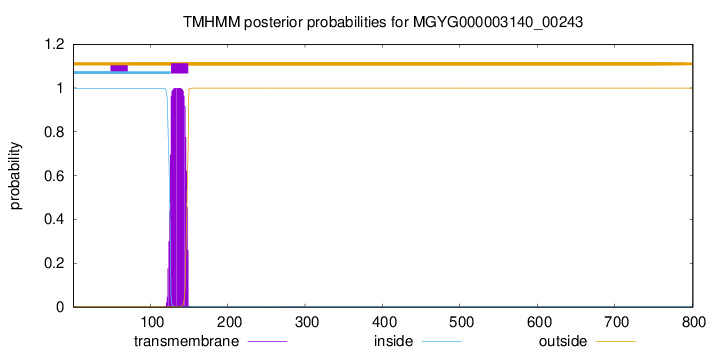
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp000758755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp000758755 | |||||||||||
| CAZyme ID | MGYG000003140_00243 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20514; End: 22919 Strand: + | |||||||||||
Full Sequence Download help
| MAPSQANDDQ GRRALPSRAE ATRRRLAQQA RREGEGTIPQ TPASSAPKRT SAVARVSRGS | 60 |
| SSGAAASSSG SRRASTRAAS SGSTSRTGST RPASSRTAAS GKGGGKKKGP AKGKKKRSLA | 120 |
| ARIARGAFYT FLGLIIAGFG TFLIAYASLK IPQADQVALA QTTTVYYADG TTEMGRMSQV | 180 |
| NRKIIDSSTL PDYVSHAVVA SEDRTFYTNS GVDLKGIGRA LVNNIRGGAR QGASTLTQQY | 240 |
| VERYYVGETT GYVGKAKEAI LALKINREQT KDEIIGNYLN TIYFGRGAYG IEAASQAYFG | 300 |
| HEAKDLTLSE AAMIAGIIPA PSAWDPAVDE EKARSRWQRV INLMVEDGWI SQADANAAEF | 360 |
| PKTIDPDTLG GPTMEGPTGY LMEQVRTELA AEGFTADQLA TGGYRIISTI DKDKQDQAVA | 420 |
| AAEYMYTVRG WNEDSMHVAL TSLDPATGEI VAEYGGKDYL TRQQNAATQD IMAAGSTFKP | 480 |
| FALVAHAEQG GSIYDVYDGS SPKTFSGMSR AVSNDSGVSW GRQNLVNATK NSINTVFVGL | 540 |
| NQEVGPATTK KVAIDLGIPE DTSGLDDSLL NVLGFAAPHN IDLAHAYSTI ASGGKRTTPH | 600 |
| IVREVQNSDG SKVFKTNVDP EQVFDTKTMS TVLPALEAVT ASGGTAEAAA ALKQDTGGKT | 660 |
| GTSEDQKTAQ FVGFTPELVT AVSMYQLDEN GNPVSLTNIG GLGQFHGGDW PVTVWMRYMK | 720 |
| QATANQTKLH FDWYTRQSAT PVAPAPQVST TPTPSEETPT PTPEATESTP GSEVQPTQTP | 780 |
| TAQNTGAAGS EGGGPSRAEG R | 801 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 173 | 345 | 2.2e-61 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 1.73e-138 | 117 | 786 | 9 | 661 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 7.33e-132 | 181 | 725 | 1 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 3.41e-101 | 128 | 794 | 9 | 795 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 2.93e-74 | 172 | 345 | 3 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| PRK09506 | mrcB | 3.89e-46 | 182 | 729 | 211 | 749 | bifunctional glycosyl transferase/transpeptidase; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYB16859.1 | 4.57e-279 | 114 | 794 | 71 | 762 |
| AOS47583.1 | 5.36e-278 | 115 | 738 | 73 | 700 |
| SDT85356.1 | 4.88e-277 | 113 | 756 | 63 | 700 |
| QWW20550.1 | 1.12e-274 | 97 | 753 | 80 | 736 |
| QCT34577.1 | 1.53e-273 | 107 | 780 | 53 | 714 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZG8_B | 2.90e-45 | 264 | 702 | 2 | 449 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3UDF_A | 3.48e-42 | 185 | 722 | 42 | 715 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 3ZG7_B | 7.94e-42 | 264 | 682 | 2 | 428 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 5FGZ_A | 1.33e-38 | 182 | 727 | 156 | 694 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 1.49e-38 | 182 | 727 | 177 | 715 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0R7G2 | 1.57e-79 | 123 | 682 | 71 | 647 | Penicillin-binding protein 1A OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=ponA1 PE=3 SV=1 |
| P71707 | 2.95e-79 | 123 | 727 | 133 | 753 | Penicillin-binding protein 1A OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ponA1 PE=1 SV=3 |
| O86088 | 2.21e-57 | 122 | 722 | 4 | 705 | Penicillin-binding protein 1A OS=Neisseria cinerea OX=483 GN=mrcA PE=3 SV=1 |
| P0A0Z6 | 3.52e-56 | 139 | 722 | 19 | 705 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup B (strain MC58) OX=122586 GN=mrcA PE=3 SV=1 |
| P0A0Z5 | 3.52e-56 | 139 | 722 | 19 | 705 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
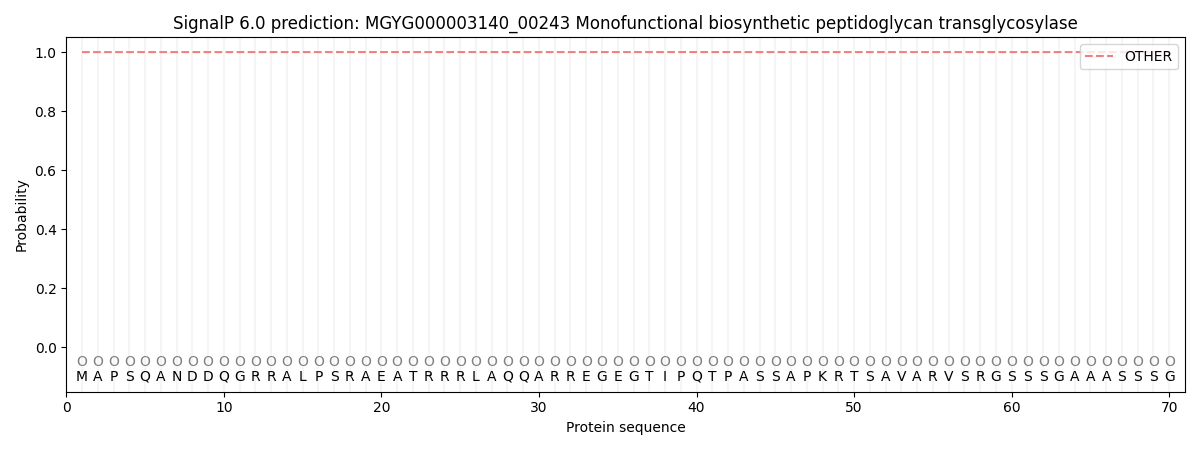
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000048 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |