You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003462_00651
You are here: Home > Sequence: MGYG000003462_00651
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
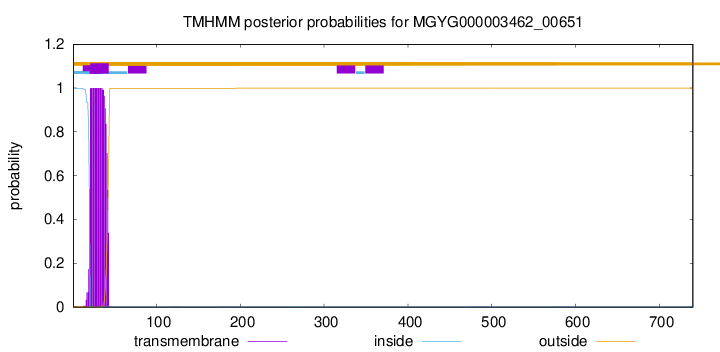
TMHMM annotations
Basic Information help
| Species | Collinsella sp900767675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900767675 | |||||||||||
| CAZyme ID | MGYG000003462_00651 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3692; End: 5914 Strand: + | |||||||||||
Full Sequence Download help
| MGPRQRQGRS RSKTHALPMI LLSFLGLLLI AGVAFGIGMI GNVNRWLSDL PDYTDANAYL | 60 |
| VSEPTEIVDC NGNKIASFYT QNRKSITKDQ VSPYVLQGTV DVEDERFYEH GGIDLWGIAR | 120 |
| AAVATLSGGH EGASTITQQL VRNTILQEEQ FDSTIERKVR EAYIAIKMEE MYSKDEILMM | 180 |
| YLNTIYYGHG AYGIEAAAET YLSKSAADLT LSEAALLIGL PNSPSQYDPT VNPDLALSRR | 240 |
| NKVLDNMLRL GHITQKEHDA AQAEPITLNV TEISGSGVDV YSQPYFVAYV KQLLEQEFST | 300 |
| DVLFKGGLTV KTTIDPTMQQ TAEEAVRAQL DTLSLDGLDM GMVVVDPKTG YIKCMVGGYD | 360 |
| YNADENHVNH ATAKRPVGST FKAFTLATAI QNGMNPNVTL NCNSPLTAKG TTTKVQNYAN | 420 |
| QSYGTITLKQ ATAYSSNTGY IQVAEAIGNN KIMSLVKKLG IDPSKDNIED VPVMTLGTGS | 480 |
| ISPLEMAAAY ATFANGGYYR QPIAITEIDS RTGSVLYQHT DNPSQVLSTG EAAAVTDVLT | 540 |
| GVMQGSGTGA AGALSVNQPY AGKTGTTDNT TNLWFCGYTP QLACALWTGY SAGEIPIQKY | 600 |
| GMDLLGDSTN LPVFKKFMNT VLAGTGREEF PTGTAPTYKN NNVWKFYGTS SARKSENEEA | 660 |
| EDKGGTTTTT TTTTTTTETT GGDTTGGNGA TGGDGTTGGD GTTGGGSGGE GGSPTPTPTP | 720 |
| TPTPTPTPTP TPTPDPSTGQ | 740 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 72 | 247 | 5e-66 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 0.0 | 82 | 624 | 1 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 0.0 | 30 | 631 | 24 | 606 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 7.24e-157 | 30 | 630 | 13 | 727 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 2.36e-84 | 71 | 248 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| PRK11636 | mrcA | 3.43e-82 | 50 | 631 | 33 | 794 | penicillin-binding protein 1a; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA33680.1 | 0.0 | 1 | 689 | 1 | 692 |
| AZH69246.1 | 0.0 | 1 | 689 | 1 | 692 |
| ATP53936.1 | 0.0 | 1 | 654 | 1 | 654 |
| AEB07395.1 | 1.42e-282 | 1 | 652 | 1 | 652 |
| QWT17307.1 | 7.07e-270 | 1 | 646 | 1 | 646 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UDF_A | 1.58e-63 | 58 | 625 | 14 | 719 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 4OON_A | 3.58e-62 | 64 | 622 | 20 | 723 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 5FGZ_A | 1.27e-58 | 103 | 601 | 176 | 678 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 1.66e-58 | 103 | 601 | 197 | 699 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3DWK_A | 1.11e-56 | 64 | 601 | 7 | 564 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70997 | 9.14e-86 | 53 | 654 | 43 | 642 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| P38050 | 1.14e-80 | 55 | 590 | 39 | 570 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| O66874 | 7.51e-79 | 33 | 630 | 13 | 676 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| O87626 | 1.56e-70 | 35 | 625 | 19 | 708 | Penicillin-binding protein 1A OS=Neisseria flavescens OX=484 GN=mrcA PE=3 SV=1 |
| A7GHV1 | 8.68e-70 | 9 | 589 | 5 | 617 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
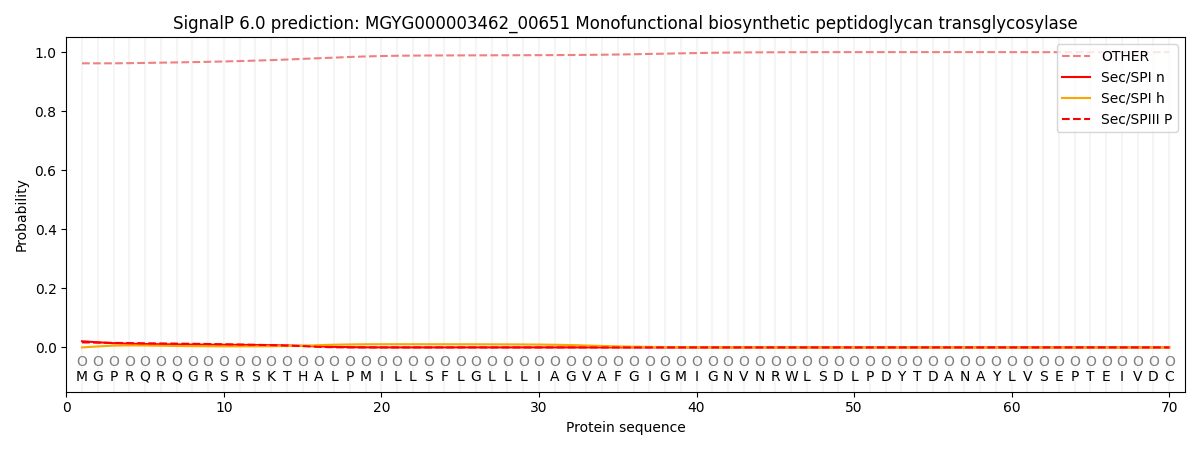
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.963158 | 0.019093 | 0.000161 | 0.000140 | 0.000068 | 0.017403 |