You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003671_00474
You are here: Home > Sequence: MGYG000003671_00474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
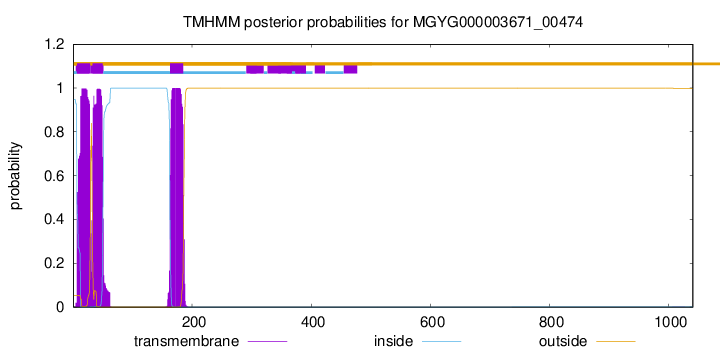
TMHMM annotations
Basic Information help
| Species | HGM10818 sp900772445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; HGM10818; HGM10818 sp900772445 | |||||||||||
| CAZyme ID | MGYG000003671_00474 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35938; End: 39063 Strand: + | |||||||||||
Full Sequence Download help
| MKDFIKKINV FDIIIGILFL VINILSFILF KWLGLLSIPI FGVLIFIYYN SKEIGNMISE | 60 |
| LINKNNKSKN KKNNKGKNNK TNNKSIKNSK NKNKNNSIDD EFDDDDDIII KKKKTENVYE | 120 |
| TKDVDMVKIN NKKKGKSKKN NKSKTKKKVN KEKKTGKQIL KSILIVVFSF CIFCVLSLAA | 180 |
| FLTYIVITTP NFDPDKLTYK DQTVVYDKNS VAFATLGTQK RESVTYDELP QVLVDAIIAT | 240 |
| EDSRFFQHNG VDFARFLKAS FEQLLHIGTG GGASTLTMQV SKNNLTNTES RGIKGIIRKF | 300 |
| RDIYISVFQI EKKYTKEQII EFYVNDNLLG GSNYGVEQAS QYYFGKSVSE LNLAEAATIA | 360 |
| GIFQSPNAYR PDRNPEGAEK RRNTVLYLMK RHGYITEEEE AITKAISVES LVVSHPENET | 420 |
| YQGFLDTVTA EVESKTGNNP YEIPMKIYTT LDTTVQNGID DIMEGKTYTW QNDVVQAGVA | 480 |
| VVDVNTGEVI AIGAGRTKDI LGGWNYATQE RNHPGSTAKP LFDYGPGFEY SNYSTYTLFN | 540 |
| DEPWSYTNGP AVNNWDSSFH GLMTLRTALS QSRNIPALKA FQQNNKKNIL DFVTKLGITP | 600 |
| ETPLHEAHAI GGFNGVTPLE LSAAYATFAN GGYYIEPHTV TKIIYRDNDK EEEFKYTKER | 660 |
| VMSASTAFLI NNVLQYAVET GYGGGARVYG KTVAAKTGTS TYGDDIIEKY GMGYDAVNDL | 720 |
| WTAAYTPQYS FALWYGYKER SSEYYNTNTT GAVYKNNLMS ALVKVIPMTD KAFDVPDTVV | 780 |
| QSQVEFGTWP AQLPSEYTPK DLINTEYFIK GTQPTETSQR FTKFTDVKNL ETTKINNNTV | 840 |
| ELTWNYDMPE IFTETYLKKY FSQNVFGNGT DSFVKNRLNY YGGVGYGIYT IDENNNLTKI | 900 |
| DFTTDKEYRY TNTTSKDVHI VVKVQYKDFD GNASNGVEND ILIRGKNSNS NSDSKLTTNI | 960 |
| NSSGTYNVGN YVESDFTIKY NGRTISKDEV TIKYSVNGVE YFDKEDLVSS INSLSSGTYT | 1020 |
| ITYTISYKDE TEKFTKRITL N | 1041 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 213 | 390 | 4.7e-56 | 0.9717514124293786 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 2.39e-155 | 155 | 822 | 8 | 657 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 8.20e-108 | 157 | 840 | 2 | 783 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| COG4953 | PbpC | 2.46e-76 | 176 | 736 | 7 | 534 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| TIGR02071 | PBP_1b | 3.30e-73 | 219 | 705 | 145 | 639 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| pfam00912 | Transgly | 6.32e-67 | 209 | 390 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNF30636.1 | 1.11e-152 | 185 | 847 | 62 | 725 |
| APH04640.1 | 1.34e-149 | 185 | 851 | 57 | 720 |
| AYA76804.1 | 1.26e-148 | 179 | 847 | 53 | 726 |
| AWI12666.1 | 3.08e-148 | 184 | 847 | 55 | 716 |
| QNG61477.1 | 3.67e-148 | 158 | 846 | 33 | 717 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DWK_A | 2.57e-83 | 205 | 739 | 10 | 559 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 6.01e-80 | 190 | 739 | 4 | 568 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
| 3ZG8_B | 1.11e-58 | 309 | 752 | 2 | 447 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3ZG7_B | 2.38e-55 | 309 | 752 | 2 | 447 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 4OON_A | 1.97e-45 | 205 | 737 | 23 | 718 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39793 | 9.65e-140 | 190 | 852 | 64 | 735 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| Q00573 | 7.00e-95 | 184 | 740 | 34 | 603 | Penicillin-binding protein 1A (Fragment) OS=Streptococcus oralis OX=1303 GN=ponA PE=3 SV=1 |
| Q04707 | 1.61e-87 | 184 | 740 | 34 | 602 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=ponA PE=1 SV=2 |
| Q8DR59 | 5.85e-87 | 184 | 740 | 34 | 602 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=pbpA PE=1 SV=1 |
| P38050 | 7.01e-73 | 176 | 800 | 22 | 632 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
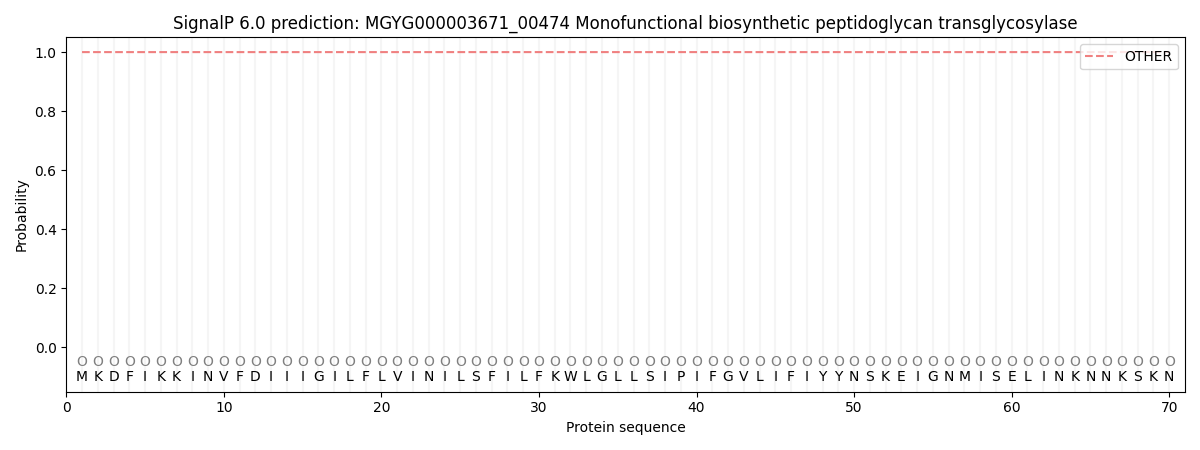
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999994 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |