You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004259_00117
You are here: Home > Sequence: MGYG000004259_00117
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
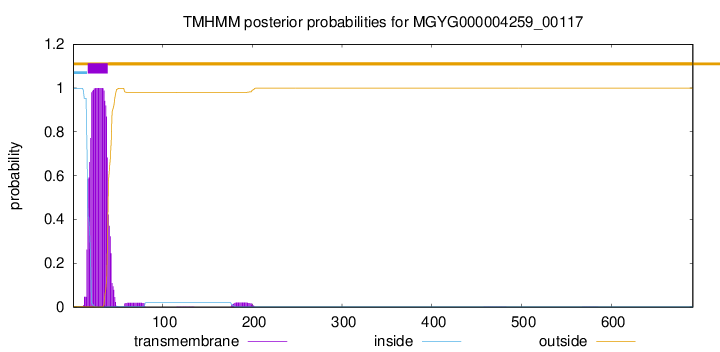
TMHMM annotations
Basic Information help
| Species | Atopobium minutum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Atopobiaceae; Atopobium; Atopobium minutum | |||||||||||
| CAZyme ID | MGYG000004259_00117 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1F | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 130256; End: 132331 Strand: + | |||||||||||
Full Sequence Download help
| MGVRTRRAHR HAQTHAFGFG VLGVFGFIGL LGIALMVSLG SLMAFWLTDL PDYKSADAYL | 60 |
| VAEPTTVYDV KGNVIANFYL QNRKSVTMED VSPYVLKGTV DTEDLRFYKH NGVDPYGIMR | 120 |
| AIFSNASGGS EGASTITQQL VRNTVLSDEQ FDKTIKRKVR EAYIAVEMEK TYTKDQILMM | 180 |
| YLNTIYYGQG AYGIEAASIT YFNKHANELT LAEAATLVGI PNSPTAYDPT VNPNDSKARR | 240 |
| NLVLKRMLKQ GDITQEEYDA ASQEDIVLNR GQSVTSEKGK YPYFTSYIAS LLSEDFDAKT | 300 |
| IYQGGMKVYT TLDPDKQEAA EKAVQAQLDS IGNKELESAL VAINNKNGYI DAMVGGRDYD | 360 |
| TNQFNLATQA RRQPGSSFKA FTLAAAISQG LNPQTYLNCN SPIQVTPTWR VQNYGNTNYG | 420 |
| TISFARATEI SSNTGYVQVA QAIGGSAISS TAKSMGIGVD IPSYASITLG TIGVPPIQMA | 480 |
| EAYSTLANNG IHRNAVAITK IEDRNNNTVY EHKEQSSQAI SGEVAYATTQ VLQGVVSNGT | 540 |
| ATAVRSYAPH VNQPIAGKTG TTENARDLWF VGYTPQTTVA VWTGFREEKT IYVHGSAGHP | 600 |
| SNTSCPILGR YLEMTLAGAE RQEFTSSPAP TYKPASSWPF AKYQKPETLA QTLEDNSQTE | 660 |
| EETDQQDTDD TEPTTSTAPD SNSSDEAAGD E | 691 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 72 | 247 | 1.7e-68 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 0.0 | 17 | 679 | 10 | 660 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 0.0 | 82 | 618 | 1 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 1.17e-169 | 28 | 625 | 11 | 728 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| PRK09506 | mrcB | 6.95e-91 | 100 | 613 | 228 | 744 | bifunctional glycosyl transferase/transpeptidase; Reviewed |
| PRK11636 | mrcA | 2.67e-89 | 30 | 585 | 16 | 743 | penicillin-binding protein 1a; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCV18421.1 | 7.84e-287 | 1 | 642 | 1 | 643 |
| ADK67697.1 | 1.16e-284 | 1 | 640 | 1 | 640 |
| SDR66234.1 | 3.68e-283 | 1 | 659 | 1 | 661 |
| BBH49739.1 | 4.03e-267 | 1 | 641 | 1 | 642 |
| QUC02420.1 | 5.32e-267 | 1 | 641 | 1 | 642 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UDF_A | 1.63e-68 | 58 | 618 | 14 | 718 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 5FGZ_A | 5.60e-68 | 102 | 612 | 175 | 690 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 7.80e-68 | 102 | 612 | 196 | 711 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 4OON_A | 5.44e-66 | 64 | 616 | 20 | 723 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3FWL_A | 1.34e-64 | 102 | 612 | 179 | 694 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O66874 | 3.65e-93 | 28 | 628 | 6 | 680 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P38050 | 3.33e-88 | 67 | 625 | 51 | 605 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| P70997 | 2.12e-86 | 53 | 591 | 43 | 587 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| O87626 | 1.42e-84 | 22 | 618 | 16 | 707 | Penicillin-binding protein 1A OS=Neisseria flavescens OX=484 GN=mrcA PE=3 SV=1 |
| O05131 | 1.99e-82 | 17 | 620 | 14 | 710 | Penicillin-binding protein 1A OS=Neisseria gonorrhoeae OX=485 GN=mrcA PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
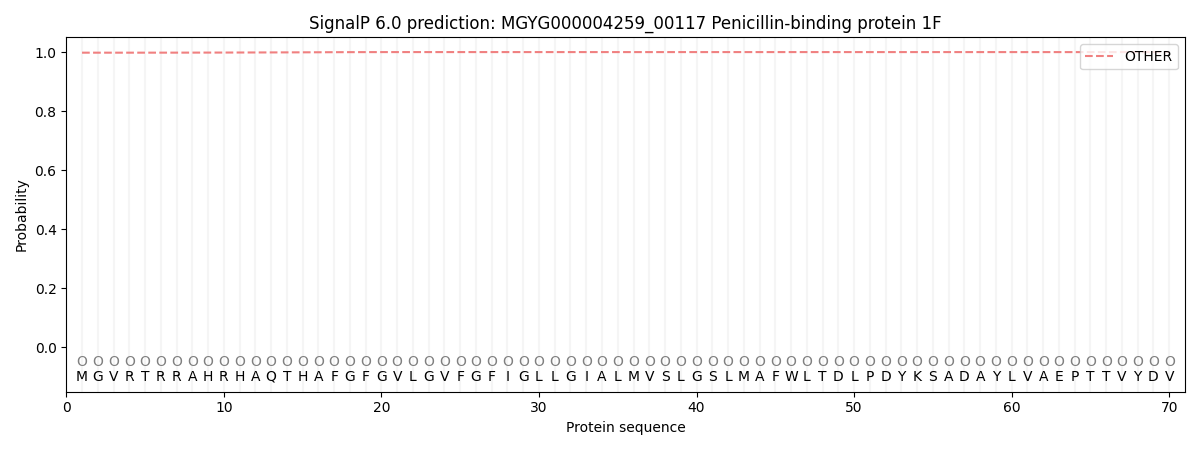
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.998328 | 0.001561 | 0.000014 | 0.000013 | 0.000006 | 0.000139 |