You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004261_00165
You are here: Home > Sequence: MGYG000004261_00165
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
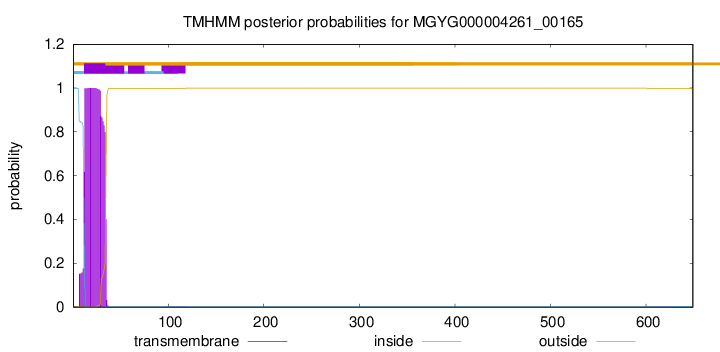
TMHMM annotations
Basic Information help
| Species | UMGS1449 sp900551925 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; UMGS1449; UMGS1449 sp900551925 | |||||||||||
| CAZyme ID | MGYG000004261_00165 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 2D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2975; End: 4924 Strand: + | |||||||||||
Full Sequence Download help
| MKKGQKIVKI GVITLLILIG IFILFNLIGF LYAYITPKLD ISSANAFSVY DNKGSLVFQG | 60 |
| NGNDSWVSLD KMNDNIINAT LSVEDKNFYR HWGFDFPRIG AAFLANISSG EIVQGASTIT | 120 |
| QQYAKNLFLD FEQSWTRKWK EMWLTYELEA HYTKDEILEG YLNSINYGHG MYGISNASKY | 180 |
| YFDKEVGELS LAEASILAGI PNSPSDYSPI DNYEMAKKRQ KVVLERMYNN GYITKKEMDI | 240 |
| AYKTKLNFKN DSFDNELTSL IYYNDAVMNE LEEIGTIPSS YLDTGNIKIY TTLDIEAQKS | 300 |
| LEAGLSEIAI NDEIETSKIM MDPDDGAIIA LIGGLNYGES QFNRVINSYR QPGSTVKPFL | 360 |
| YYRALENGFT PSTTFFSKET SFYFDNKETY TPANSGNIYA NKEISLAAAM AYSDNIYAVK | 420 |
| THLFLGEFEL YNTLRKVGIT SKMEKSPSLT LGTYEVSIIE LARAYSALAN SGHKVEPHLI | 480 |
| TKVVDKNDNV LYEYKEKEEE VVLNSDITFM ISELLTTTYD TNLIDYTYPT CINMVDRITN | 540 |
| KYAIKSGSTD TDAWVIGYTP EVVFASWAGY DDNKEISTEV VSANKDSWIT SMEKYFETND | 600 |
| ATWYDVPNNV VGVLVNPITG NVVSNSNEKK KILYYLKGTE PYIKEDSDD | 649 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 61 | 228 | 2.9e-66 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 1.11e-171 | 66 | 597 | 4 | 530 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG0744 | MrcB | 5.79e-166 | 27 | 573 | 29 | 574 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 8.32e-133 | 27 | 646 | 22 | 769 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 5.60e-83 | 63 | 228 | 12 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG4953 | PbpC | 1.28e-71 | 46 | 574 | 36 | 539 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYF83258.1 | 6.34e-176 | 2 | 641 | 10 | 653 |
| QNK49558.1 | 5.07e-175 | 2 | 641 | 10 | 653 |
| QOR66557.1 | 1.08e-174 | 17 | 641 | 24 | 653 |
| AZV45469.1 | 1.26e-174 | 2 | 641 | 10 | 653 |
| AZV62027.1 | 2.03e-174 | 2 | 641 | 10 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZG8_B | 1.96e-58 | 147 | 587 | 2 | 451 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3UDF_A | 8.57e-56 | 75 | 603 | 50 | 723 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 3ZG7_B | 1.29e-55 | 147 | 587 | 2 | 451 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 4OON_A | 4.51e-55 | 49 | 620 | 23 | 747 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3DWK_A | 8.56e-55 | 49 | 575 | 10 | 562 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70997 | 7.17e-153 | 49 | 641 | 57 | 653 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| O66874 | 1.30e-82 | 5 | 643 | 1 | 714 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P38050 | 1.09e-69 | 67 | 571 | 70 | 571 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| P39793 | 6.43e-67 | 2 | 641 | 23 | 691 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| Q68VU2 | 1.12e-65 | 66 | 623 | 69 | 744 | Penicillin-binding protein 1A OS=Rickettsia typhi (strain ATCC VR-144 / Wilmington) OX=257363 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
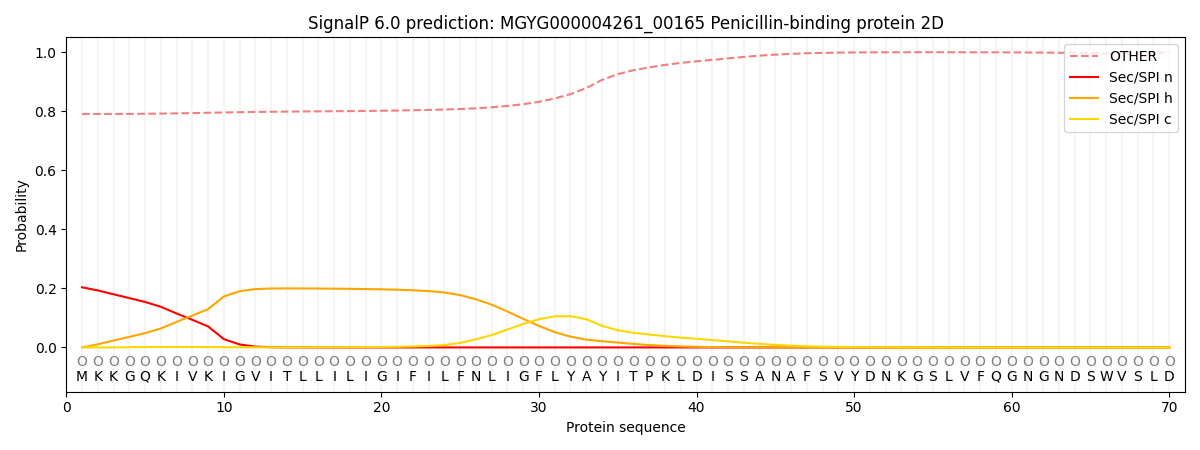
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.800022 | 0.190311 | 0.001049 | 0.000937 | 0.000485 | 0.007219 |