You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004546_00802
You are here: Home > Sequence: MGYG000004546_00802
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
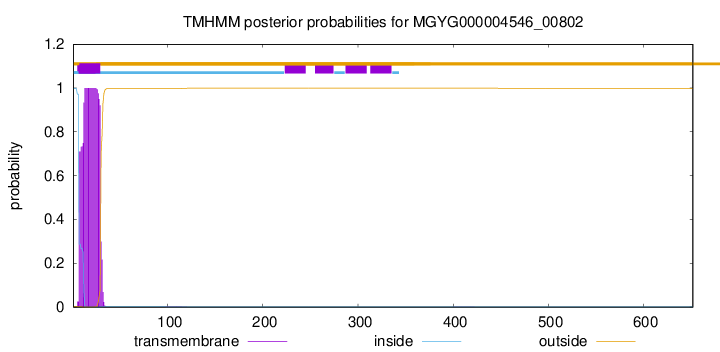
TMHMM annotations
Basic Information help
| Species | CAG-460 sp900751815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-460; CAG-460 sp900751815 | |||||||||||
| CAZyme ID | MGYG000004546_00802 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 2D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 96; End: 2054 Strand: + | |||||||||||
Full Sequence Download help
| MKKVRFLFKL MLFGFISFIV IIIGLYTYAY LSPKLDIKTS GSLYLYDSNN ELVYQGSRNN | 60 |
| EWANLEDISD YLIDATIAVE DKNFYKHHGF DYLRIIKAMY LNIKSGTIVQ GASTISQQYI | 120 |
| KNLYLDFDQT WERKIEEAFL TLELEVHYEK DEILEGYLNT INYGQGNMGI VSAAEYYFNK | 180 |
| KPSELSLEEA IMLAGISKNP ARYNPVSDYD ACIARAKVVA KSMLNNEYID EDTYNSLFQD | 240 |
| KIEIYGQNKT NNLQMLMYYQ DAVMDELASI SEIPESLIDA GGLKIYTTLD IDMQANLEEN | 300 |
| ILENKPDDEI QVASVVVDPN TGAIRALTGG MDYAKSQYNR ALESKRQVGS TMKPFLYYAA | 360 |
| LENNMTMSST FSSVPTTFTL SNGQTYSPQN ADSLYADKEI TMAAALALSD NIYAVKTNLF | 420 |
| LGVDKMIEVA HRTGIEADLD EVASLPLGTS EINLLDFATG YTTFATGGYK KDLYFIERIE | 480 |
| DLEGNVLYEK EHEKKLVLNP NYTYILNEMM NTTSNDAYVD YTTPTALTVS SGFTNKYAIK | 540 |
| TGSTNTDFWI VGYNKDALVM TWMGYDDNKA VTSRVRTSSK KAWTKTIEYA LKDIDTSWYE | 600 |
| TPENVVAIPL DAVTGKVTND NNKVALYYYV KGSEPSVLNE QYVSSENEKN TE | 652 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 56 | 223 | 5e-62 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 3.44e-176 | 1 | 630 | 9 | 630 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 1.61e-173 | 61 | 593 | 3 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 1.19e-131 | 1 | 636 | 2 | 765 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02071 | PBP_1b | 1.94e-100 | 65 | 568 | 152 | 660 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| PRK11636 | mrcA | 8.09e-84 | 1 | 636 | 1 | 829 | penicillin-binding protein 1a; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMC08260.1 | 1.83e-165 | 20 | 630 | 29 | 644 |
| QQX26484.1 | 1.81e-164 | 2 | 647 | 11 | 665 |
| QJS18329.1 | 5.93e-162 | 19 | 636 | 23 | 646 |
| QQZ09247.1 | 7.25e-161 | 19 | 647 | 31 | 665 |
| QJX64342.1 | 3.53e-158 | 2 | 636 | 13 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FGZ_A | 6.90e-62 | 70 | 569 | 166 | 672 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
| 3VMA_A | 9.20e-62 | 70 | 569 | 187 | 693 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli K-12] |
| 3FWL_A | 5.39e-58 | 70 | 569 | 170 | 676 | CrystalStructure of the Full-Length Transglycosylase PBP1b from Escherichia coli [Escherichia coli] |
| 3UDF_A | 7.20e-57 | 43 | 598 | 21 | 723 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 4OON_A | 3.90e-56 | 67 | 640 | 46 | 773 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70997 | 1.35e-134 | 32 | 636 | 46 | 654 | Penicillin-binding protein 2D OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpG PE=2 SV=3 |
| O66874 | 2.61e-67 | 56 | 637 | 59 | 714 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P39793 | 3.63e-67 | 32 | 649 | 63 | 705 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| O87626 | 7.55e-66 | 33 | 606 | 36 | 720 | Penicillin-binding protein 1A OS=Neisseria flavescens OX=484 GN=mrcA PE=3 SV=1 |
| O87579 | 1.98e-65 | 56 | 607 | 64 | 722 | Penicillin-binding protein 1A OS=Neisseria lactamica OX=486 GN=mrcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
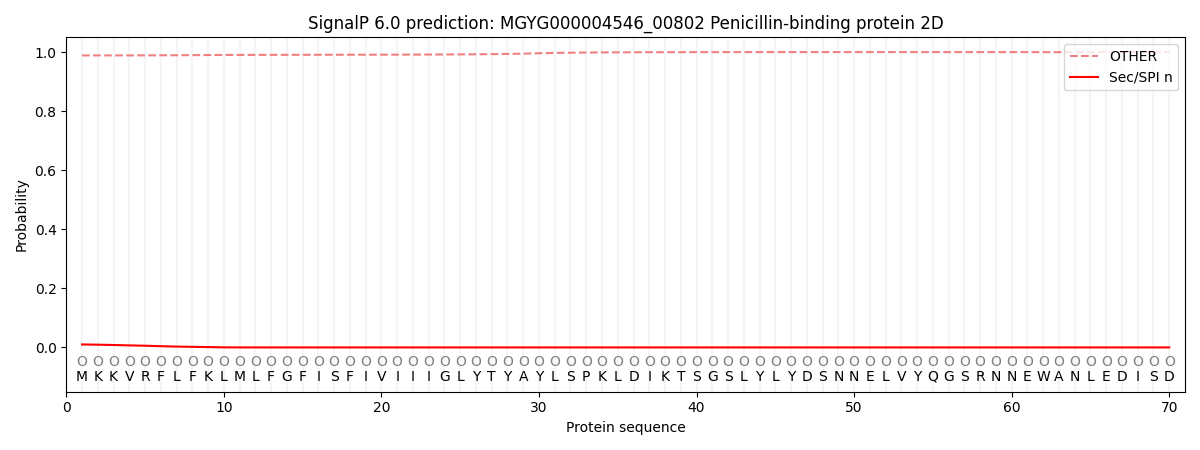
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.989240 | 0.009261 | 0.000222 | 0.000084 | 0.000039 | 0.001182 |