You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004628_00598
You are here: Home > Sequence: MGYG000004628_00598
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
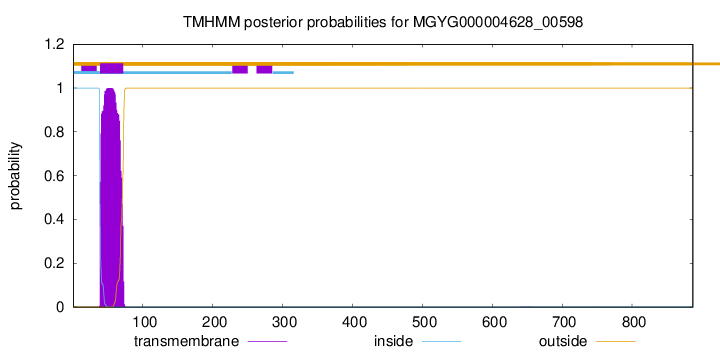
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Lawsonibacter; | |||||||||||
| CAZyme ID | MGYG000004628_00598 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61754; End: 64417 Strand: - | |||||||||||
Full Sequence Download help
| MSEQNYSSRY PSRDERQSRS GQNGEAPRRR RKKSKKKKGG VLRVIGTLLL IFLTTSAFLA | 60 |
| CFAAVYIVNV ILPQANMDLS SFNVDENSVM YYQDKSTGEY KELRQVLSVT DSQWVDYEDM | 120 |
| PQYLKDAAVA IEDRRFWTHN GVDWGRTAKA VISMFTGGDI QGGSTITQQL IKNLTDYNET | 180 |
| TVKRKVTEIF RALYVDANYE KEDILELYLN VIPLGSGCEG VGAAAEKYFG KSVSDLTLAE | 240 |
| CASLVGITNN PSKYGPYSTA RVANSDGEMW DAVQWNKYRQ EVILGQMLEQ GKISQAEYDE | 300 |
| AVAQELVFVG SDADNGQSAS NIYSWYEEQV ITDVMNDLKE EFGYSSQYVS QMLSNGGLRI | 360 |
| YTCVDPEVQA AAEAIYYDRS NLNYTSSKSG QQLQSAITII DNTTGDIAGL VGRIGEKTIN | 420 |
| RGTNLATGAL RQPGSSIKPL TVYAPAMDMG LLTPISVLAD YPYQVLNGKA WPVNVDGRYR | 480 |
| GQVTVTEALQ WSYNTVAVRT VALVTPTKSF EFATQRFHLT LESGRMVNGE MQSDVNLAPL | 540 |
| AMGGLTDGVT TRQMANAYAV FPENGIYREA RTYTKVEDSD GNVILDNSRE DELATKESTA | 600 |
| YYMNTMLQNV VQSGGGTEAQ LSGMTVAGKT GTTDSKYDRW FVGYTPYYTA AVWVGYEYNE | 660 |
| RVPASNNPAA QLWKKVMAPI HEGLENKSFD KPSGLVKVSY CLDCGGIATS ACQYDPRGSR | 720 |
| VATGYVMQED APCTSCTCHS LEPGSNSLVQ VCVDSPILDE NGQPTGGYHL AGPYCPQESI | 780 |
| RSYAYLNLDR ESVGGAWAED SGYFYSAQAD PGVCTVHTGE AVVPPEEGET TNPDDPSDPN | 840 |
| NPTDPNNPTD PNNPTDPNNP SQGGDTEGGG TQEGNGETTE QPSNPTN | 887 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 108 | 287 | 3.5e-59 | 0.943502824858757 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 3.32e-168 | 41 | 692 | 11 | 608 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| TIGR02074 | PBP_1a_fam | 8.51e-168 | 113 | 683 | 3 | 531 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 1.03e-115 | 42 | 698 | 2 | 736 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| TIGR02071 | PBP_1b | 5.16e-82 | 118 | 678 | 153 | 680 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG4953 | PbpC | 4.39e-74 | 114 | 714 | 60 | 594 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA31344.1 | 2.08e-271 | 43 | 834 | 32 | 821 |
| ANU41033.1 | 1.06e-270 | 43 | 834 | 8 | 797 |
| QBB65500.1 | 8.31e-238 | 43 | 837 | 3 | 795 |
| ALP94484.1 | 1.65e-237 | 20 | 837 | 9 | 827 |
| QNL43378.1 | 1.31e-208 | 41 | 818 | 26 | 777 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 1.45e-76 | 115 | 713 | 43 | 794 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 3ZG8_B | 7.90e-64 | 195 | 661 | 3 | 437 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3ZG7_B | 2.26e-57 | 195 | 661 | 3 | 437 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 2JE5_A | 3.09e-53 | 115 | 735 | 57 | 696 | StructuralAnd Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6],2JE5_B Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6] |
| 3UDF_A | 4.81e-50 | 115 | 657 | 42 | 689 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TNZ8 | 6.48e-107 | 65 | 740 | 48 | 725 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=pbpA PE=3 SV=1 |
| Q0SRL7 | 1.00e-106 | 66 | 740 | 49 | 725 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain SM101 / Type A) OX=289380 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 9.45e-100 | 85 | 732 | 54 | 695 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A7FY32 | 2.71e-99 | 85 | 732 | 54 | 695 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 2.71e-99 | 85 | 732 | 54 | 695 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
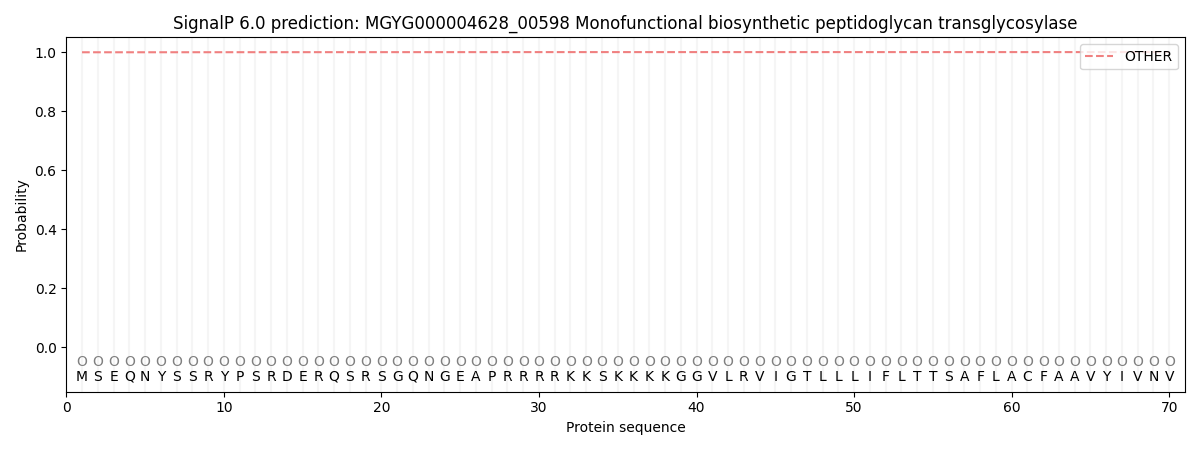
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999628 | 0.000370 | 0.000001 | 0.000000 | 0.000000 | 0.000013 |