You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000619_00342
You are here: Home > Sequence: MGYG000000619_00342
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900540625 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900540625 | |||||||||||
| CAZyme ID | MGYG000000619_00342 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75625; End: 77232 Strand: + | |||||||||||
Full Sequence Download help
| MKRNTIILII IAGALLLPWL GLPWFNSKGE PREAIVAVSM LNSGDWILPT SCGGDIPYKP | 60 |
| PLLAWLIAVF SEIFNGGVVS TWTARLPSAL SAIFLVIFGW KVMAPRIGRD RAWMTFGLTL | 120 |
| TSFELFRTAI ACRVDMVLTA CIVGACYSIF LMYETDGRRR LWHALAAILL LSGGILAKGP | 180 |
| VGALLPLLAM GIYRLICRDN VLRTIGVMLG LLMASLLLPA LWYYAAYLRG GDEFVRLALE | 240 |
| ENIGRLTGTM SYDSHVNPWW YNFATLLWGV LPWTIPALMA LCLRRVRTAI ATVWCNRDRK | 300 |
| PDLTKMAWTV ALTVLIFYCI PSSKRSVYLL PMYPFIAMGL SWLLWQCRTA KFMRIFARVL | 360 |
| ALLAIIAPVA LTLLSHISSS HLRIYTPAWW QYIMAFLPVI AGVWWLVTRS REANAAAGAV | 420 |
| VIAYLLFMSY ASTFAPMLLN PKSDRPAAEV VARQVPKDAP LYGVITGDSL LRYYTINYYL | 480 |
| DDRMNHATDI NAVPRGAYVV TDPQAVDNRP DYTVLDTLTS RSCDTRTATV LAHRK | 535 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 3 | 475 | 1.8e-72 | 0.8629629629629629 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.07e-25 | 30 | 500 | 35 | 485 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 5.56e-07 | 59 | 196 | 2 | 131 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 4.77e-05 | 30 | 426 | 33 | 424 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAJ74247.1 | 4.61e-29 | 5 | 344 | 12 | 339 |
| ACN14933.1 | 5.19e-28 | 6 | 338 | 13 | 332 |
| VDY09698.1 | 1.04e-27 | 10 | 474 | 22 | 491 |
| VDY15546.1 | 1.04e-27 | 10 | 474 | 22 | 491 |
| CDW96010.1 | 1.04e-27 | 10 | 474 | 22 | 491 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 1.10e-15 | 7 | 474 | 34 | 496 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8FRR0 | 1.97e-11 | 16 | 331 | 21 | 319 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
| A4SQX1 | 8.01e-11 | 27 | 341 | 27 | 327 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas salmonicida (strain A449) OX=382245 GN=arnT PE=3 SV=1 |
| A0KGY4 | 3.25e-10 | 27 | 338 | 27 | 324 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1 |
| B4ETL9 | 1.75e-09 | 30 | 397 | 36 | 391 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT2 PE=3 SV=1 |
| C5BDQ8 | 1.40e-06 | 30 | 331 | 32 | 319 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999820 | 0.000166 | 0.000037 | 0.000000 | 0.000000 | 0.000003 |
TMHMM Annotations download full data without filtering help
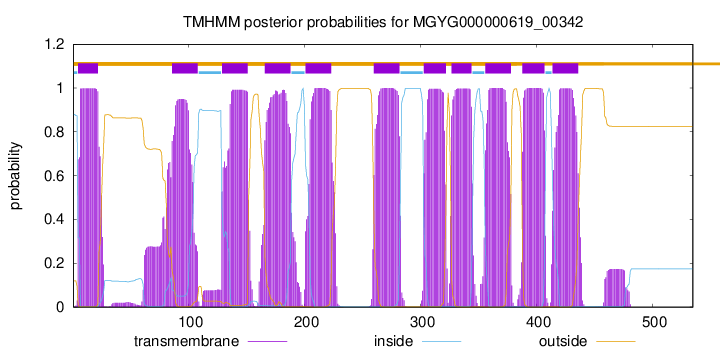
| start | end |
|---|---|
| 5 | 22 |
| 86 | 108 |
| 129 | 151 |
| 166 | 188 |
| 201 | 223 |
| 260 | 282 |
| 303 | 322 |
| 327 | 344 |
| 356 | 378 |
| 388 | 407 |
| 414 | 436 |
