You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001324_00151
You are here: Home > Sequence: MGYG000001324_00151
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusobacterium periodonticum_D | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium periodonticum_D | |||||||||||
| CAZyme ID | MGYG000001324_00151 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 163512; End: 165071 Strand: - | |||||||||||
Full Sequence Download help
| MFSTRRKDIF VLVVLSLFAY LSIIAIREID SAEARNFIAA REMLENSSWW SPTVNGHFYF | 60 |
| ENPPLPTWLT AIVMMITRSH SEAILRIPNM LCCIFTILFL YRSMIRIKKD RLFAFLCSFV | 120 |
| LLTTFMFIKL GAENTWDIYT YTFAFCASLA FYLYVRDGQR KNLYRMAILV FLSFLSKGPI | 180 |
| GFYSVFIPFL LAHYIIFPKE IFKKRTFFVF LTLIISIALS LVWAFSMYFN HGDYFLTIVK | 240 |
| DEVSAWATKH HRSFIFYTDY FVYMGSWLFF SIFVIFKVPE KKEEKVFWLW TILSLIFISI | 300 |
| IQMKKKRYGL PMYLTSSITI GQLCIYYFRK TYAELKKREK TLLIIQQLFL LFVIFVSLIF | 360 |
| LTYFGYIKKE ISFGLFFLYA ALHLLFLFLF AVGYTEISYA KRVIIFSGLT MLLVNFSSSW | 420 |
| ILESKFMQNN LLKFRMPIDE EILKSSDSIY AERYDIEDVW KLGKQIKTLN KNMPDEREII | 480 |
| FLGKEEPKSL SKVYEVKKVY EYQKVTHDME RLYILERIY | 519 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 4 | 431 | 2.2e-52 | 0.8111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 7.49e-57 | 1 | 519 | 2 | 529 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 3.65e-11 | 63 | 219 | 3 | 153 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279 | arnT | 4.18e-07 | 4 | 224 | 3 | 224 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1928 | PMT1 | 0.002 | 47 | 230 | 56 | 269 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASK79558.1 | 2.08e-15 | 33 | 454 | 40 | 466 |
| CAB1276937.1 | 7.74e-13 | 5 | 311 | 8 | 326 |
| ADR19199.1 | 9.91e-13 | 3 | 345 | 3 | 349 |
| QQB17357.1 | 1.36e-12 | 30 | 311 | 31 | 326 |
| SNV00837.1 | 1.36e-12 | 30 | 311 | 31 | 326 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.994830 | 0.004817 | 0.000072 | 0.000036 | 0.000027 | 0.000241 |
TMHMM Annotations download full data without filtering help
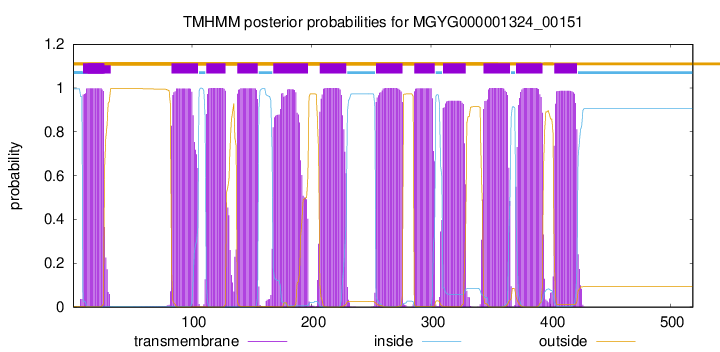
| start | end |
|---|---|
| 9 | 26 |
| 83 | 105 |
| 112 | 128 |
| 138 | 155 |
| 168 | 197 |
| 207 | 229 |
| 254 | 276 |
| 286 | 303 |
| 310 | 329 |
| 344 | 366 |
| 371 | 393 |
| 403 | 422 |
