You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000732_00358
You are here: Home > Sequence: MGYG000000732_00358
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-245; | |||||||||||
| CAZyme ID | MGYG000000732_00358 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 99222; End: 107543 Strand: + | |||||||||||
Full Sequence Download help
| MKEKRKLSIK GAILDKNQLK DYLEKIASDH ILNEKSDRDT YPIPRLKENF FVIKEIYKLL | 60 |
| NEQIKQGIPI HPAGEWILDN LYVIEEIVKN IIKELTLKKY TDFLGLANGR FKGFARVYVL | 120 |
| ATEMVAFTDG KINSENLEYM LQAYQTKKTL SMNEIWNIGL FIQIALIENI REICETIYMS | 180 |
| QMQKSRVEEI LGKYFGEEKK KIRPILVQFV HFGDGEKNAL IEYMSYRLKK MGSKAYAYLN | 240 |
| ILEEEVAKTG SDIYEVVKKE HFDIAVKKVS VGNCISTLKA ITRINFLEIF EKINRVEDIL | 300 |
| KKDPAKQYEK MDSTTKSYYR NAIQEISKKT KISEIYIAKK CLELSLQASE KPEANEKSMH | 360 |
| IGFYLISDGK ETLLNSLLEK KITLKSNEEK SKIYIETVWG ISILLTLILC INFYYMLNIE | 420 |
| NWMRVLLSII TAIVAILPIE NIVSKIIQYV LSKVVKPKLI PKIDFQNGIP EDCSTMVVIP | 480 |
| TIVKSKEKVK ELMRKLEVYY IANKSSNLYF TLLGDCCSGS KEVEEFDEEV IREGIEQSKR | 540 |
| LNEKYGNIFN FVYRKRIWNP NEECYMGWER KRGLLNQLNE YLLGNIANPF RANTIDISQM | 600 |
| NKIKYIITLD SDTDLTLKSG LELVGAMAHI LNKPEVNEKC DLVISGHALM QPRVGVGLVE | 660 |
| SRKSIFTQVY AGEGGTDSYT NVISNLYQDN FDEGIFTGKG IYDLSIFSKV LANEIKENTV | 720 |
| LSHDLLEGSY LRCALTSDVM LMDGYPSNYM SFRTRLYRWI RGDYQILPWL GKTIENKKGE | 780 |
| IKQNPLKLLS KYKIFSNIVR SKQESSVLAM LVFSVAIATL LKVNMCGIIV LALISSLIPT | 840 |
| ILDIINSIIS KKDGNIKTKR FVKTIDGLKA SVLRGVLDIG LIPDKAWITT KAETKTIYRM | 900 |
| TKSKKHLLEW ITSEEAERTA KTDIFSYYKN MNANELIGLL CVIFAILAYI QNCSTGIAIF | 960 |
| TLTIGLYWLL TPALMCKISK ANKEIKAVEK IDESEKKYLQ EVAYRTWLYF KENLTQKSNF | 1020 |
| LPPDNYQEDR KPKVVMRTSS TNIGLALLAV MASYDLRFEN LEDTLNLLYK MLETISGLTK | 1080 |
| WNGHLYNWYN IETLEPLIPK YVSSVDSGNF VGYMYVLKQF LIEKSVHNGD GEKNVQTTSK | 1140 |
| VHNEEGKKDV QITLEMLNKI DFMINATDTL INNTDFSKLY DNENRLFSIG FNVEENKLTD | 1200 |
| SYYDLLASEA RQTSLVAIAK KDVTEKHWYN LSRTLTTLNN YKGLISWSGT SFEYLMPTIN | 1260 |
| IEQYPGSILD ESCKFMIMSQ LEYAKKLGIP WGISEAAFNL KDLNNNYQYK AFGIPWLGLK | 1320 |
| RGLSDEIVTS SYGSILAIDE EPKLVIENLK NLSEKGMYNK YGFYESIDFT PGRAKNGYTP | 1380 |
| VKTYMAHHQG LILLSIDNLL NNDVIKKRFK QNTEIEAVDI LLQEKMPENM ITTKEEKEKI | 1440 |
| EKIKYVDYED YTQRKYSRIN ENLNVSNVIA NDNYTIVLDQ YGNGYSKYGD LQVNRYKETD | 1500 |
| EAEQGIKFYI KNIRNKNIWT NTYSKNLRIP DKYNIIFSPE ANKIVRNDEN IRTVTKIIVD | 1560 |
| TDDPVEIRRL ELKNNGVSEE VLEITALLEP VLSNAMQDFA HKAFNNLFIS FEYIDSINTI | 1620 |
| VVKRKAHTNG EKDVYMAVNL FSQSNELGDV EFEIDKEKLW GRCNYRIPRE VENSIPFSKK | 1680 |
| IGYTTDPIIA LKKTVDIKPE ETAVFDLIVS VGYDKEQVIK NMAKFMNNEN TKRTFELLKA | 1740 |
| KSEAENRYLG IKGKDIEVYQ KMLSYLLFTP KVSIKALYDI PCPVSELWKY GISGDLPILM | 1800 |
| VKIKDINDID IIQEVLSAYE FFRVKNIKVD LVILNEEKES YERYVKDAIQ RAIFNRNLAY | 1860 |
| LLNIPSGIFC LENVEKKDKK LLEERANLVI NAYYSSLALQ MEDIESKMLD NVKENVFDAK | 1920 |
| QNNFIEVNDE KENLIDESSL KYFNEYGGFS RDGKEYLICI NKTHKLPTVW SHVLANKTFG | 1980 |
| TLTTENMGGY TWYKNSRLYR ITAWSNDQVL DTPSEIIYAK DMDTGRKWSL GFNPMPDNND | 2040 |
| YFVVYGFGYA KYIHSSSRIR QTVDMFVPEN DNIKVNLITL ENKNPQRKNL KLIYYIKPVL | 2100 |
| GEDEIKSNGY LQLEMNIASN IMLVKNLSDE KNNNQIFIGC SEKISSYTGS KTSFLKNSTL | 2160 |
| ENPVGLDELA LNRENSFGED GIVAIQINVT IEAYSTKEIV FTLGDASNRE EYQDLAYKYS | 2220 |
| NVNNCKNEYI NIRRHWEQLV NKLQINTPVE STNILLNGWL IYQTISSRMY GKTGFYQSGG | 2280 |
| AYGFRDQLQD CMLIKYVEPN IARDQILRNC RHQFIEGDVE HWWHEETDKG IRTRISDDLL | 2340 |
| WLPYVVADYI SFTGDYEILE EKLSYKDGLR LAENENERYD LYKDADFKES VYKHCIRAIE | 2400 |
| KAIGIEDNEV EEIKNAENNE TPRITIRNFG KHGLPKIGTG DWNDGMSNVG AKGKGESVWL | 2460 |
| GFFMYAVLKA FIPICERMEN LEKKEKYQLV MDGLKRALNN NAWDGRWYKR AFTDDGDAIG | 2520 |
| SLQNEECKID SISQSWATIS GAGNNDKKYI AMESLENHLV DKEIGIIKLL DPPFEKSKLE | 2580 |
| PGYIKAYLPG TRENGGQYTH AATWTIIAQT MLNLNDKAYE NFRMINPIEH ARTKEESSKY | 2640 |
| KVEPYVIAAD VYGQGNLAGR GGWTWYTGSS SWMYIAGIKY ILGLNIESGY LKIEPHIPQN | 2700 |
| WDNYEIKYKY KNSVLNIKIT RGSLDKKSEK MVENNTNISD TTIVKQMICN GEEIPEKQIK | 2760 |
| IGENGVYNIE VII | 2773 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1652 | 2403 | 2.3e-183 | 0.6795366795366795 |
| GT84 | 1201 | 1412 | 2.9e-85 | 0.9906976744186047 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1651 | 2773 | 1 | 1052 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 3.63e-130 | 2235 | 2684 | 1 | 424 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 2.67e-80 | 1940 | 2220 | 2 | 284 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 9.39e-76 | 1967 | 2204 | 1 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| pfam06165 | Glyco_transf_36 | 3.87e-63 | 1466 | 1708 | 5 | 245 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV69563.1 | 0.0 | 6 | 2764 | 31 | 2909 |
| AUG57227.1 | 0.0 | 7 | 2772 | 32 | 2908 |
| ADU74104.1 | 0.0 | 5 | 2710 | 34 | 2870 |
| ANV75789.1 | 0.0 | 5 | 2710 | 34 | 2870 |
| ABN52453.1 | 0.0 | 5 | 2710 | 34 | 2870 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 3.10e-84 | 1945 | 2719 | 2 | 770 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 6.00e-66 | 1945 | 2719 | 2 | 761 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 5.56e-59 | 1946 | 2743 | 3 | 807 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 2CQS_A | 3.08e-58 | 1946 | 2720 | 23 | 804 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3RRS_A | 3.19e-58 | 1946 | 2743 | 3 | 807 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 45 | 2739 | 61 | 2823 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 8.94e-78 | 1945 | 2772 | 2 | 811 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q9F8X1 | 1.83e-68 | 1945 | 2719 | 2 | 761 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q76IQ9 | 2.99e-65 | 1945 | 2719 | 2 | 761 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 3.16e-44 | 2013 | 2773 | 83 | 790 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000068 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
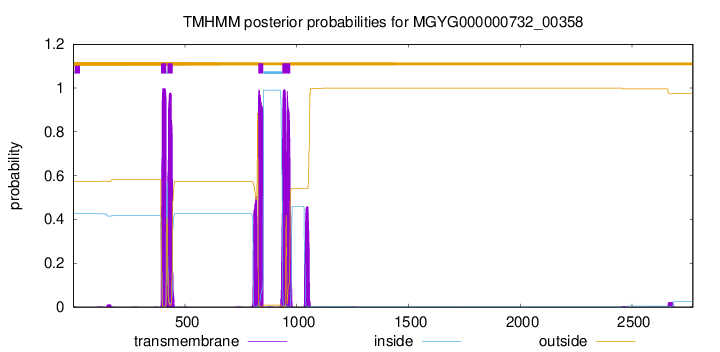
| start | end |
|---|---|
| 393 | 415 |
| 422 | 444 |
| 828 | 850 |
| 936 | 970 |
