You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001962_00004
You are here: Home > Sequence: MGYG000001962_00004
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-877 sp900554315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-877; CAG-877 sp900554315 | |||||||||||
| CAZyme ID | MGYG000001962_00004 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 492; End: 8156 Strand: + | |||||||||||
Full Sequence Download help
| MANLVKNFLS DFEDINEYYM FLVDKTKQKE YVGITNEWLI DNFYLLVEHK TNIMHDKKEI | 60 |
| TKKLKKYDRI FYCLKDIVSK NNYNISFKLL VTELKDYQKQ TKVYFSYPEL VGVKNVLLFI | 120 |
| YTKRLADLCH EEYKNLVNKE KVARIIASRE EKDIELSNFL SDHISIQEEA NYIFEINNQL | 180 |
| KELGAKSNRL FKELNELLEE KRISLKEIIN NEYQKSIDND ILISNIFGDL KEFFEFTEED | 240 |
| LFKKVSKTEK LLLEDEVYSK MTVESKSLYR QQLLKLAKKH HKDELSYLQE LLDNADGDEY | 300 |
| HIGFQLFPRK KNSLRVILYI LVIVLATFGI SFVLSEFFLP WRWLSFILLL IPVSQLFVQI | 360 |
| FNQILIRFVP TYPLPKLDYS KGIPDDSKTM VVIPTIISSK EKVKNMFDTL ETFYLMNKTD | 420 |
| NLYFTLLGDV TASKEEVIDL DQELTEYGIS YAEKLNKKYK KDLFFFLYRK RFWNEHENTY | 480 |
| LGYERKRGAL LQFNQLLLKK MSKKDQDYWY FANTLGDFSE DIRYVITLDQ DNRLVLNSAL | 540 |
| NLVGAMAHPM NRPVLNKEGT KVIKGYALMQ PRVSLDIEAT NKSLYSQIFA GIGGFDTYSA | 600 |
| IVPNVYQDAF GEGSFIGKGI YDVRVFDQVL SDVFPDNLIL SHDLLEGNYL RCAYVSDIEL | 660 |
| IDDFPSKFLT DTSRHHRWAR GDVQIIGWLL PFVRNKQGKK VKNPINLLGK WKIFDNIVRM | 720 |
| FLYPTLLLVL LFAIFFGKTN PLWWIGLVLL EIAIPIVFFL QSKVYQRENK KEKATVYYKH | 780 |
| LLFGGKSLVL RSYIVLSTLP YYSKLYLDAF FRTCYRLLIS HNNLLNWVTA EDAAKKVDKD | 840 |
| FVSYLRHFTF HLVFSFVLLL IGLITFNVYA CLLAIVFITS PFVLYYVSQD IDPQRKELKE | 900 |
| KEVEEVREMA YRTWCYFKDN LKEEYHYLIP DNYQENREPK LDLRASPTGI GFSLLSVISA | 960 |
| VELEFITYEE GKDLISHILD TVDSLEKWHG HLYNWYDINS LHVMHPSFVS TIDSGNFVAS | 1020 |
| LVVVREFLKN HEEVEKRKLC DKLIRNANFK KLYTKRDVFS IGYDELEGKL SIYNYNKFAS | 1080 |
| ESRLTSYLTI CFGDAPSKHW FCLDKSLTTY KGRKGLISWS GTSFEYFMPF LFMKNYPNTL | 1140 |
| LDESYHFAKF CQQDYIESVS RSLPWGISES AYNELDNSLN YKYRAFSTPY LKAKEDKENR | 1200 |
| IVIAPYASLM AMALFPRDVY ENFDKFKKLD MIGEYGFYES YDYDNKGVVK AFFAHHQGMS | 1260 |
| LVGLANYLKP GVIQDYFHQN VNIKTYDILL KEKVQVRTSI DMKMAKYKKY DYHKEIIEND | 1320 |
| IRTFNYISYL PEVSVLSNKK YCLLMNDRGD GFSRYRTLQL NRYRKVTEQD YGVFLYIKDL | 1380 |
| DTNYIWSNTY APMNKKPDKY EVVFAADKIK YLRTDGKITT KTEIVVTKNH HAEIRKITFK | 1440 |
| NESDDVKRLE LTTYTEPILS ESMDDISHRV FNSMFLTSMY ESDSNSLVVR RKSRNDTNIN | 1500 |
| SYMVHRLLIE DPMSEYSYET ERKNFLGRNQ SMQDPRAMHV SLSNEAGTNL DPVLSIRNTI | 1560 |
| EIEPNSSETV YIINGFGRSM EQIHDIISSY HDEYSIEKAF KVSTLMNVIN TKNMNITGKD | 1620 |
| MRIFNIMLDY LYQTTRLSVT EERMDLLRKN ALNQSGLWKF GVSGDRPIVL VEISDIADLT | 1680 |
| FVYDVLKAFE YYKNNSIFID VVIINSESSQ YAKIIKKEID DELYRMYTLN SFYHIPGSVT | 1740 |
| VVDGTDLTRE EKSLLYMVAR LAFPIRDHHS LQEEVEELQR NNKVSDYSKD EVEEHTELST | 1800 |
| KDRLTFDNGY GGFKNHGTEY VIYNPNTPAP WSNIIANKNF GTIITNNGCG YTYAYNSGEF | 1860 |
| KITSWTNDMV ANDKSEGFKF NGKIFDPMKC THGFGYSILE SENDELKKEI TEFVAKEDTV | 1920 |
| KFYLVKLTNK LKKAVDVDID FWINPVFGNF EEKTARHILT EFMGDDNYLK LRNVYSLTYA | 1980 |
| DVCVFMSTDL KIDSTVNDKI LVKSIHTKLH LNPKEDNTCV FVLGSSMGGD TIGDMLAKFA | 2040 |
| DVKKAKSELK AVKDYWKKTL NVVHVNSPDK SFNYMVNGWY LYQALASRIL ARAGFYQVSG | 2100 |
| AFGYRDQLQD SMNISLILPD QARRQILINA SHQFEQGDVL HWWHEKNRFG LRSRYKDDYL | 2160 |
| WLVYATVRYL DVTNDYSILS AEVPYVVGPE LSNYEHEKGM VFNYTDYKET LLDHCLKSLH | 2220 |
| LAMDSLGSHG LPLMGGGDWN DGMNKVGIKG KGESVWLGFF LYEVIDKFMA FMKTYDKKFK | 2280 |
| LKEFKDFNEK LKDNLNKKAW DGDYYLRAYF DNGDKLGSHE NKECKIDLIS QSFSILSGVA | 2340 |
| PKDRIQKVIT AVEENLVDDD SKIVKLLTPP FEKSLNNPGY IMNYPKGLRE NGGQYTHATS | 2400 |
| WYFMALIQTG YYDRAFRYYQ MINPVNRSLD VDAVNKYVVE PYVVAADIYS AERYPGRGGW | 2460 |
| TWYTGTAAWY YYVGIEEIVG IRKHGDTLTI KPNIPIAWDS FTVDYTYMDT IYHVEVHKTK | 2520 |
| KESVTLDGKK CESGIVNLVN DQKEHKIVVH YIAG | 2554 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1518 | 2549 | 1.4e-297 | 0.9980694980694981 |
| GT84 | 1075 | 1280 | 5.7e-67 | 0.9813953488372092 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1518 | 2549 | 2 | 1051 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 2.35e-120 | 2055 | 2482 | 1 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 3.54e-82 | 1273 | 1600 | 1 | 334 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 1.07e-65 | 1330 | 1573 | 2 | 245 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 1.59e-52 | 1804 | 2039 | 2 | 283 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK11488.1 | 0.0 | 1 | 2549 | 51 | 2843 |
| ADL68114.1 | 0.0 | 37 | 2549 | 102 | 2864 |
| QSZ28298.1 | 0.0 | 37 | 2549 | 99 | 2862 |
| AST58113.1 | 0.0 | 37 | 2549 | 102 | 2864 |
| QCX34529.1 | 0.0 | 2 | 2547 | 62 | 2706 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 4.33e-75 | 1810 | 2549 | 3 | 808 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 7.60e-65 | 1810 | 2535 | 3 | 785 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 6.25e-61 | 1810 | 2536 | 3 | 807 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 3.60e-60 | 1810 | 2536 | 3 | 807 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 2CQS_A | 3.79e-56 | 1810 | 2528 | 23 | 819 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 172 | 2521 | 234 | 2810 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 6.44e-73 | 1810 | 2549 | 3 | 810 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q76IQ9 | 2.82e-64 | 1810 | 2549 | 3 | 799 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q9F8X1 | 2.40e-61 | 1810 | 2542 | 3 | 787 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 5.48e-38 | 2004 | 2526 | 260 | 763 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
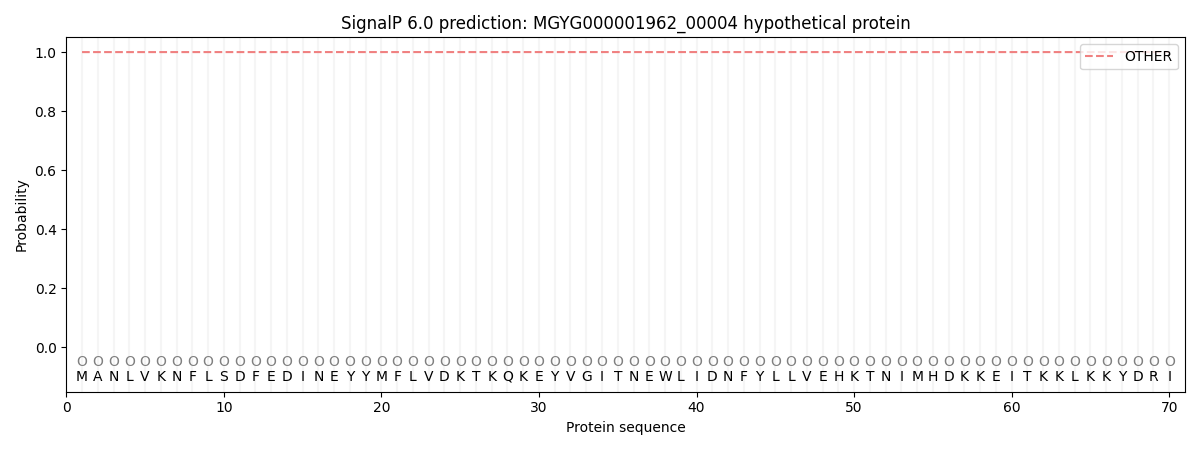
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
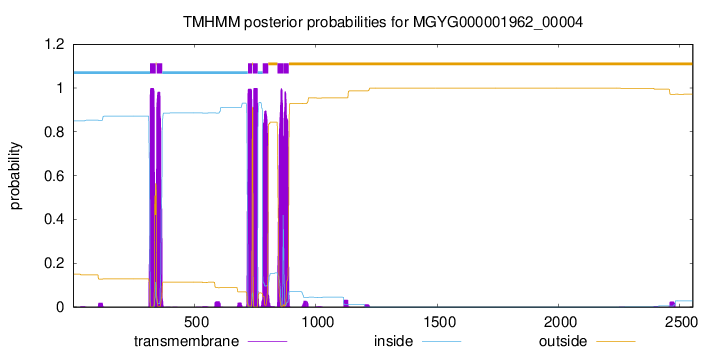
| start | end |
|---|---|
| 317 | 339 |
| 344 | 366 |
| 720 | 737 |
| 741 | 760 |
| 781 | 803 |
| 842 | 864 |
| 866 | 888 |
