You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002723_00437
You are here: Home > Sequence: MGYG000002723_00437
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS911 sp900545935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; UMGS911; UMGS911 sp900545935 | |||||||||||
| CAZyme ID | MGYG000002723_00437 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 108785; End: 116035 Strand: + | |||||||||||
Full Sequence Download help
| MRKSITRRQI HSLRARLRAN GDTTALGELE LLLALDKSGR RASGSLLGAL GEILEEKNYR | 60 |
| AVAEEIITEA EKRYGAIWEE EDVRLLPSAL LSLSLAHIAV EGSAEAISAA RSMQNSFSAI | 120 |
| SPYTASQRLS AMHKLLSEIS DEYRVSSRET QQMYRTQLVR AARRERISST EAARRYADSL | 180 |
| DTLPEVLLGC APTLYFAVYA ALTILPPAVL LPLLLMLGSG GAVLPLSLAW GSTEYIIAAV | 240 |
| ICALLLLFPS AECARLLTDR IFSHCIGYSV LPRTDDENIP DSGKTLVCIT SLLFGEERDG | 300 |
| ELFDRLERYY LGNRDENLRF ALLADLPDSD SEHEDGDEKV LEYAESKIRE LCDKYGERFF | 360 |
| LFVRSRRQAP DGRWCGWERK RGAVLELIRF CRGDAGSFSA VPEDCEFVSS ARYLITLDAD | 420 |
| TTLPIGAAHR LITAMLHPSA HPIIEDGHVV RGHSIIQPRM VTTARSASVT PFAVMCSGGG | 480 |
| GADTYASAGY DTFQSMYDEG NFCGKGIIDI DTYNKLLCDR FPEETVLSHD LLEGIILRSA | 540 |
| IDGECRFSDS TPKNPVSFMT RAHRWIRGDV QAWLWGRRED MDALSHFKLL DNVRHALTPL | 600 |
| ASLILVILSA FMGGAAAMLI VPLALSYAIL PFIITLLDRM ASLPSALAHG GAVTVRSGRI | 660 |
| RGAVMLELRR MIYSVCTLAY QAARAGDAAV RALWRTLVSH RRMLEWTTAA EGDRYSGQLT | 720 |
| DYTVRMMPSV LLGVAFMLTG SAMLRLFGLM MFIAPTLAWR LGTPYGKHCD APSIEERAQL | 780 |
| LTYAADIWRF FADTVTAEEN YLPPDNISYQ REVRVADRTS PTNIGLYLMS VIAARDLGLI | 840 |
| GTAETERRLR ATADTLVRLE RWHGHLYNWY SVSTASVLSP YVSAVDSGNF VASLMAVCSA | 900 |
| LDEYIPECPG LAEVREALER LFRGADFSVF FSPERGLMHT GINASSGELD RGFYDLYMSE | 960 |
| SRTSYYLAVA LGQLPPRAWS NLGRVYASNH AGVGMLSWSG TAFEYFMPTL YYLPAEDTAE | 1020 |
| RQSLRFGARG QRIGAYRGVW GKSEGCYCAY DRDGNYRYKA IGVSSLALGP CGGERVYSPY | 1080 |
| SSYLVMQCSR RAALENLSRL REYGMYGRYG FYESLDLTPS RVGDGHAVVR SYMSHHMGMS | 1140 |
| LAAIDNVCSG GIFRERMSRI PELACAETLT DERIPVGSLA PRVESIKTRV RRGSRTRECL | 1200 |
| PELPAGIRRA SLVSDGAMHI CLCSDGCAEL RYGRLPLCGT VSVRNDISLA RISGGADSGI | 1260 |
| MDEKKSGVDI DTESMLRPED ERTGCLRVVL RTGSDAPQIM CGSVHCDGES AELTTDSGDK | 1320 |
| LRMIPDGSDN TVLLLGSTAG ERHCSALLYL EPRLTSEDNW NSHPAYAGLG FSARFDRARQ | 1380 |
| MLIYERSDRN GGHIYLCAAP IFGRIDDFTT RRRGLFPERY TDSDIAALLG RELGGCEGVC | 1440 |
| ISPSLAMRCE SVGSFAFIIA VGRCEEQAVT AIEDKRRLLD TLGRRLPPMR DKSPTVGDRL | 1500 |
| LEELVTAAVY GREKPPARLG DAYPINELWK YGISGDVRIG VFFLSGDGEE SGKGLRELMQ | 1560 |
| CAVRLYLHGF SLDLVFAYEG SGEYYDRRRD MLLRAAESAG ADFLIGAKSG IHLVPLESGD | 1620 |
| DSAKRLLSLY SVFTLAVSDS DTAAGMTERL AAQRIPEFLP HGERRENVKV YADNSSVTAP | 1680 |
| GSGWRFGTDG GFNMRKKSSP VPWSYPLGGC CLGTLVTDRS FGFTWLSNSR ELRVTPWSGD | 1740 |
| ESGGLRGEDI VAEIDGRTYS LSLCAEKVCY RGACAVYSGS LPGHDWMVEI CCEARLPVKL | 1800 |
| YRISGLDCCE LRLSLCGREG AVAHEHMSDC DLYTVLAGAL AGRTVAVLRP CPGVFALTVF | 1860 |
| STPLGSHSAA ELGSFPLLRY LREHYSGEGA VCDVFDRRDI GSGISMRGCF GEIDALLGSF | 1920 |
| LLHQTYYSRM RGRTGFAQPG GAYGFRDQLQ DAGALVYADT RLARCHIIRC ASRQYREGDV | 1980 |
| MHWWHDVPLP HGGISIRGVR TRCSDDRFWL PFIIGDYISV TGDAAVLDVP VPYLVSAPLA | 2040 |
| DREDDRYEDA VFSEERESIY QHCMRAMRTI RLGLYGLPLI GSCDWNDGLS GIGRNGGVSV | 2100 |
| WLGFFLRTVI GRMLPICERM GEYDDIAFLA SQYKKLTAAT LDPRHRCGDY FIRAYYGDGV | 2160 |
| PLGCENGSEP VLDSIAQSWA YFAASERVDV EKYLASGKPT SEVLDSISNE LCYARGAVRL | 2220 |
| AYQRLFDRSV GTAALLSSPL RRSDGHDPGY ICSYPEGMRE NGGQYTHAAV WLARALIRGG | 2280 |
| AAEEGMALVR ALDPSSHRTE IYLGEPYAIA GDVCTAGELR GMAGWTQYTG ASGWYYRLLL | 2340 |
| EDMLGYRECG DSFTLEPCPE LPNRLELKIE RKATVYRITF VSGTSAGIPA SEPTHKKAAR | 2400 |
| FPYDGKEHTL TYYFGT | 2416 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1913 | 2407 | 2.7e-117 | 0.4314671814671815 |
| GT84 | 953 | 1160 | 1.8e-55 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 1.71e-98 | 1914 | 2387 | 582 | 1024 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 2.38e-66 | 1913 | 2347 | 18 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| pfam10091 | Glycoamylase | 5.87e-11 | 952 | 1161 | 1 | 215 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 1.13e-04 | 1713 | 1740 | 42 | 69 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam11329 | DUF3131 | 0.001 | 782 | 894 | 1 | 115 | Protein of unknown function (DUF3131). This bacterial family of proteins has no known function. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDZ23197.1 | 0.0 | 244 | 2408 | 302 | 2491 |
| BCI59942.1 | 2.70e-314 | 245 | 2357 | 300 | 2420 |
| ADU26013.1 | 1.37e-301 | 243 | 2410 | 298 | 2537 |
| AVQ95160.1 | 1.68e-301 | 243 | 2410 | 308 | 2547 |
| AYF37850.1 | 1.68e-301 | 243 | 2410 | 308 | 2547 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2CQS_A | 1.59e-23 | 1940 | 2391 | 381 | 814 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3AFJ_A | 3.62e-23 | 1940 | 2391 | 381 | 814 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
| 3ACT_A | 1.08e-22 | 1940 | 2391 | 381 | 814 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127],3ACT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127] |
| 3S4C_A | 1.80e-22 | 1940 | 2387 | 361 | 790 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3ACS_A | 2.45e-22 | 1940 | 2391 | 381 | 814 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127],3ACS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 2.76e-212 | 207 | 2389 | 416 | 2814 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 4.07e-24 | 1944 | 2379 | 359 | 771 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q9F8X1 | 8.49e-21 | 1913 | 2388 | 313 | 771 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q76IQ9 | 6.73e-19 | 1913 | 2358 | 313 | 738 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q92AT0 | 2.46e-06 | 1940 | 2388 | 604 | 1041 | 1,2-beta-oligoglucan phosphorylase OS=Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) OX=272626 GN=lin1839 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
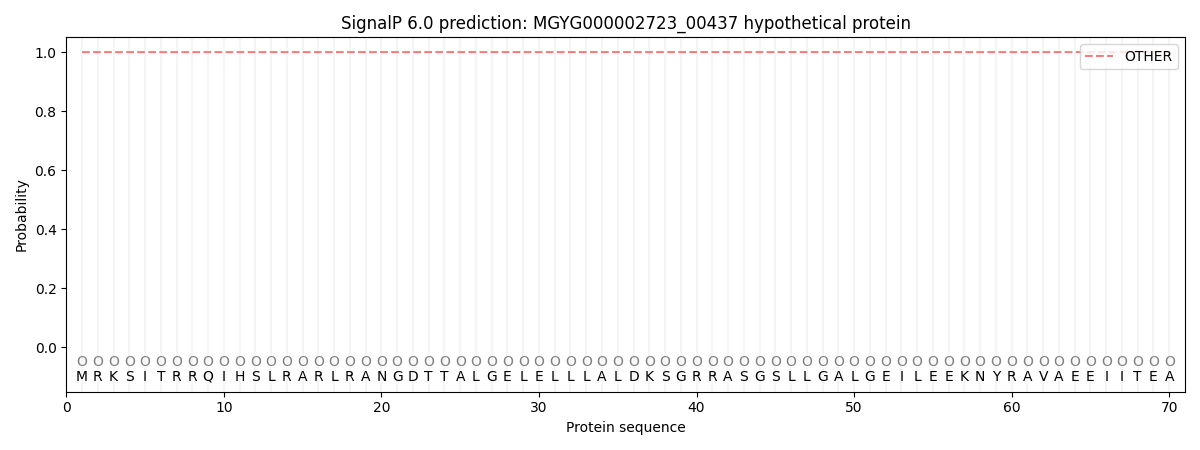
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000037 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
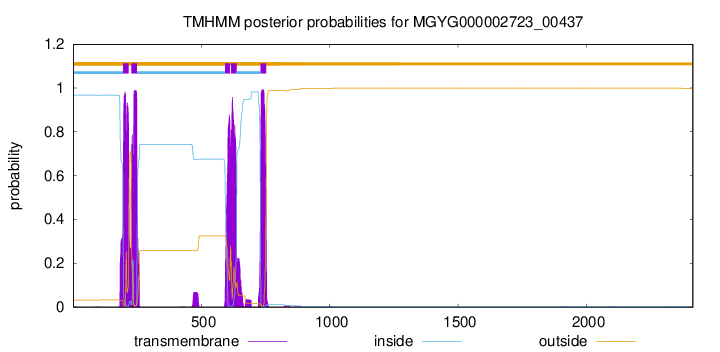
| start | end |
|---|---|
| 194 | 216 |
| 226 | 248 |
| 592 | 611 |
| 615 | 637 |
| 730 | 752 |
