You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003086_00335
You are here: Home > Sequence: MGYG000003086_00335
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-269 sp900554365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-269; CAG-269 sp900554365 | |||||||||||
| CAZyme ID | MGYG000003086_00335 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 57192; End: 65273 Strand: + | |||||||||||
Full Sequence Download help
| MKVLKINGTP LDKLQLETHL QKIASNHILI TKSQKNTYPI PHLVESFKVI QEVYNLLNEH | 60 |
| LKLGISIHPA GEWLLDNFYI IEECVKQVQK SLTLEKYKNF LGLATGEYKG FARIYILAAE | 120 |
| IVAYTDNKID RKDLEQYLAS YQTKKTLSME EIWNIGIFLE IAIIENIKKV CEKIYISQIE | 180 |
| KYKAESIVER LVENKPKQEQ KYNMNSTKKI EKIIFNNMQY SFVEYMSYIL KRYGKKGYSY | 240 |
| LKALEEIVEL TGTTVSDVIK KEHFDIAVSK VSIGNSITSI KKIQRINFLE IFEKINGVEE | 300 |
| LLKQDPSGIY EKMDNKTKES YRNKIKEISK KTKISEIYIA KKLLEIAGRC LDLESKEAHI | 360 |
| GYYLIDKGID ELYQALNFKT KKAVGSKNKA KIYITTVVVL DLIISCYFGM LLNKGINKTW | 420 |
| IFALSTILLF IPSSEIVIQL ISYISSKFVK PKLIPKMDFS SGIPKKESTM VVIPTMLKSK | 480 |
| EKVQELMKKL EVFYLANKSE NLYFCLLGDC SQSKVKEEAF DSEIISEGEK LTEELNKKYK | 540 |
| TEEIPIFNFI YREREWNKSE KSYLGWERKR GMLMQFNEYL LGNSKNIFKA NTLEKCEKEK | 600 |
| LKEIKYIITL DADTDLVLNS AFELVGAMSH ILNKPVIVDN KVVYGYGIMQ PRIGINLNIS | 660 |
| YYNLFTKIFA GDGGIDCYTN AISDIYQDNF EEGIFTGKGI YDLKVFSRIL KDKIPENTVL | 720 |
| SHDLLEGNYL RCGLVSDIIL MDGYPTKYNS FMNRLARWIR GDWQITRWLL PKYGLNLISR | 780 |
| YKILDNLRRS LLEISVIISM LFIASINKSY LWILLVPITP FILEFLNIFI FRKEGEEKQK | 840 |
| SFTPKISGIK GSFLRGILTL GCLPYKAYIS LKSILITLYR KNISHKHLLE WTTSEEAEKQ | 900 |
| AKSDLISYQK QMIINIIFAL VFILYSIYKN QIYGIIFPIL WMITPIFMWF ISKPKNESKA | 960 |
| IDKLNKEERE YVKEIGNKTW NFFKTYLNKE TNYLIPDNYQ EDRREKVILR TSSTNIGLSL | 1020 |
| IAVVSAYDLK YIDLENCLDL LENMIYSIID LPKWNGHLYN WYQIKTKEPL IPRYISTVDS | 1080 |
| GNFVGYLYVI KSFLTEAIYK ITDEEKNFKI QSLIKDVEKL INETDFSVLY NYELQMFSIG | 1140 |
| FNIEENKLTD SYYDLLASEA RQASLVAIAK KDVPSKHWNY LSRTLTILGH YKGLISWSGT | 1200 |
| AFEYLMPDIN IPRYEGSLLD ESCKFMIMSQ RKYAKELNIP WGISEAAFNV KDLKSNYQYK | 1260 |
| AFGIPWLGLK RGLADELVVS TYGGVLAIND VPKEEVENLK ILEKEGMYDK FGFYESIDFT | 1320 |
| PERVEKGQNS SVVKTYMAHH QALILLSINN LFNNKILQKR FMENPEIEAV SILLQETMPE | 1380 |
| TSIITKEKKE KVEKLKYKDY ENYIKTTYSK IDERIITGNV ISNENYVIAM NQKGEGISKY | 1440 |
| KDIYINRYKY TDDYSQGIFF VMKNIKTKQL WSSNYNFNNK KEENYQISFT PDKMEQEILN | 1500 |
| GNIKTVINTT VTSNEPVELR QVTLENRGND EEIIEITSYF EPVLSKKEQE YAHPAFNKLF | 1560 |
| LVTNYDDDTN SLIIKRKKRE QNSQDMYAFV NLSTNGETVG DLEYEINESK FTGRGNINIP | 1620 |
| NMIANSIPFS KKIGLVTEPI IAMKRTIKIK PQEKVDVNFI ISVEENCQIA KENLIKYKSF | 1680 |
| ENVKMAFELS KARVEAESRY LRIKGKEIAL YQKILSYIVF ENQVKSTRIK KINLAQYNQS | 1740 |
| ELWKYGISGD LPIILVKIKD VNDSYVIKQV LKAYEFFKTK NIQIELVILD EEKHSYDNYV | 1800 |
| KEEIESAILN QHMAYLKNRK GGIFSLVKSE IDKKDISLLE LLAIIIVDSS KGGLENNIKD | 1860 |
| IEEDYLEKNK VQETEVFPTS FIEENDEDID VLSDSENLKY CNEYGGFSND GKEYLIRVNK | 1920 |
| NNRLPTVWAH ILSNEKFGTV ITENMGGYTW YKNSRLNRVT SWENEPNYDI PSEIIYLVDK | 1980 |
| ESGKAWSLGL NPMPDDRNYN VVYGFGYAKY IHKSNGIEQE MEVFVPEEDS CKISILNLKN | 2040 |
| VTPNKKKIKL YYYTKQVLGE DEIKTNGYIN TSFDKNNNFI YAKSIYKTDE QNYFSYISCN | 2100 |
| EKIKSYTGDK KSFIGDGDIG KPESIKKTKL NDDSGLGKDS CMAMEIEIEL ESYANKDIIF | 2160 |
| MLGAEVNLID GKNTAYKYSK ISNCKQELNK VKNKWKELLG RLQVYTPLES LNILLNGWVG | 2220 |
| YQTISSRLYA RSGYYQSGGA YGFRDQLQDT LGIKYLDTNI MKNQIIKHSR HQFIEGDVEH | 2280 |
| WWHEETGRGI RTRFSDDLLW LVYLTIEYIK FTGDKSILNI ETPYLEGKLL ENNQDERYDK | 2340 |
| YETSKIEGTI YEHCIKSIEK GLNFGENGLP KIGSGDWNDS FSTVGNKGKG ESIWVGFFLY | 2400 |
| TILEQFIPIC IERNDEILAQ KYEEIKQNLK KALNTNGWDG RWYKRAYMDD GNILGSMQND | 2460 |
| ECRIDSIAQS WSVISNAGDN DKKYIAMESL ENHLVDRENG IIKLLDPPFE KGKLEPGYIK | 2520 |
| SYLPGVRENG GQYTHAAVWV IIAESLLGFG NKALELYRMI NPIEHTRTKE ACNKYKVEPY | 2580 |
| VIPADIYGAG NLIGRGGWTW YTGSSSWYYE AGIKYILGLI IEEGYMKFNP CIPKDWKNYK | 2640 |
| IQYKWKESVY NIEVKNLTGK EKEVNKIILD GNIVENKIKL DGSRKIYNIE VIM | 2693 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1603 | 2690 | 0 | 0.9980694980694981 |
| GT84 | 1151 | 1365 | 4.4e-88 | 0.9953488372093023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1603 | 2692 | 1 | 1051 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 1.53e-131 | 2195 | 2619 | 2 | 424 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 3.26e-89 | 1898 | 2167 | 2 | 272 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 6.37e-79 | 1925 | 2163 | 1 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.06e-63 | 1357 | 1689 | 1 | 336 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUG57227.1 | 0.0 | 2 | 2693 | 30 | 2909 |
| AEV69563.1 | 0.0 | 4 | 2693 | 32 | 2919 |
| ADU74104.1 | 0.0 | 4 | 2693 | 36 | 2919 |
| ANV75789.1 | 0.0 | 4 | 2693 | 36 | 2919 |
| ALX08042.1 | 0.0 | 4 | 2693 | 36 | 2919 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 2.35e-101 | 1904 | 2693 | 3 | 810 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 6.87e-87 | 1904 | 2678 | 3 | 785 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 2.13e-72 | 1904 | 2678 | 3 | 806 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 3.05e-71 | 1904 | 2678 | 3 | 806 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 3AFJ_A | 9.44e-67 | 1904 | 2656 | 23 | 805 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 45 | 2654 | 64 | 2805 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 1.55e-96 | 1904 | 2693 | 3 | 812 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q9F8X1 | 5.94e-89 | 1904 | 2675 | 3 | 782 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q76IQ9 | 3.32e-86 | 1904 | 2678 | 3 | 785 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 4.99e-41 | 1971 | 2664 | 83 | 761 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000070 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
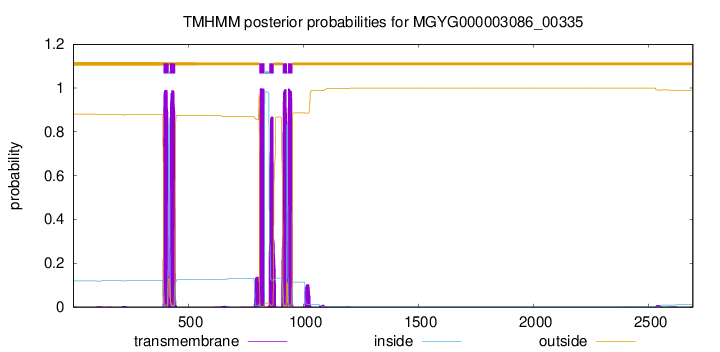
TMHMM Annotations download full data without filtering help
| start | end |
|---|---|
| 392 | 414 |
| 421 | 443 |
| 809 | 831 |
| 852 | 871 |
| 911 | 928 |
| 933 | 952 |
