You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003139_00319
You are here: Home > Sequence: MGYG000003139_00319
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_G cochlearium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_G; Clostridium_G cochlearium | |||||||||||
| CAZyme ID | MGYG000003139_00319 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | Cyclic beta-(1,2)-glucan synthase NdvB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83253; End: 91730 Strand: + | |||||||||||
Full Sequence Download help
| MDYLKFEEEC PNNTTVFRCK YKLKNTLDKS YKDILKGYIY LNDEFKNRGE IIPAGDWLLD | 60 |
| NLYLIEKEYK NIKNDVREMY YRNIPNIKSG YMKCYPRIYY VAKKIVKQNN GKLTKENIEK | 120 |
| YIEDFQKHTV LTSSELWALP TMLKIACINS TSKIVKEMVY AQKEKNRAKK LVETLLQNEN | 180 |
| NENAINGAIR RDEDFSLYFV EALIKGIRNN SIENQTIYNW INEKLDMEDK NIKDIILVEH | 240 |
| KNQAFFKMIL GNNITSIREV GIINWNTSFE KLSKLDYILN TDPANVYKSM DFKSKDYYRN | 300 |
| KIERIAKKTN LGEGFIARKA IECAKENVEQ RDKTYLNHVG YYIIDNGIGC LKEKLKHKDI | 360 |
| GISKIDSLNL DNKTNYYTWF IRITTIILSI LIAYYNLYKN YTPLWKFIVG VLILIIPMSE | 420 |
| IVISILNWIL NNYLEPDFIP KIEFKKDIPK EYSTMVVIPT LVNSKKRVSE LMEDLEVYYL | 480 |
| ANRSENIYYG ILADFKDSDK KEEEGEAEIN KFALEETKRL NKKYSKNGKN IFYFFNRYRQ | 540 |
| FNEKQGVWLG WERKRGKLEE FNRLIRGDKE TSYNVISGDI KNLYEVKYII TLDADTQLPM | 600 |
| GMAKKLIGAM AHVLNTPYID YKNKKVLRGY GLMQPRIGVG VLSGNKTLFS KIFSGETGID | 660 |
| IYTCAVSDIY QDLFGEGIFT GKGIYDVDIF NYMLKDEIPE NSVLSHDLLE GSYVRTALVT | 720 |
| DLELIDGYPA YYNASCKRLH RWVRGDWQLL PWIFKKNSLN KLSIWKMKDN LRRSLLTPSI | 780 |
| VVLIILSLIL FPKPEMWLSI AIISFLTPVL FNISEITNTS LKDISIIAKL ENWKMSLIQF | 840 |
| FLMFSFIPYK SYVMIDAISR SLYRMFISKK NLLEWQTAAD VEAKSGKSIK DYIKTMWIGS | 900 |
| FIAVLVSLIA FYKSTNLGLL LFPSCIIWFF SPCIAYYISK DIIEDTYELN KDEKMLLRRF | 960 |
| SRKIWAYFED FAEEESNWLA PDNYQEEPQK GVAYRTSPTN IGMGLIACIS ALDLGYINLK | 1020 |
| QCIYKVENII KNMNDLERYE GHFYNWYDTK SKKPLNPKYI STVDSGNLIG YLWLLENTFY | 1080 |
| EFLEEPFINI KDISKGIIDL LYLSMEEIGE EKQNYIKSKE ILDTFEGDVF SFIKILKELK | 1140 |
| LESYDCIKGK ENLYWNTKLY KTLKIFEEDF NTIFPWSQLI EESKLYETLD NNIIKELSNL | 1200 |
| ITKCSLQNFE DAILKIGDYL KNSKEEYLDK LNINIQNSKN EIKNLKDKIV NIIKNLNSIS | 1260 |
| DKMNFKMLFD EGRGLFAIGY DVEKHCLDNC YYDLLASEAR QASFITIAKG DIPYTHWFNL | 1320 |
| GRSISAIGRK KGLVSWSGTM FEYLMPLLIM KSYPNTLLKE TYDFVVESQK KYGEKKNVPW | 1380 |
| GISESGFYAF DVNLNYQYKA FGVPGIGLKR GLETELVISP YSTFMALNVD YKSAIDNLKR | 1440 |
| IIDIGAEGRY GLYEAIDFTK NRNPKDSNFK IVKSFMVHHQ GMSFMALNNI LNNFILQRRF | 1500 |
| HRDNRVKSVE LLLQEKIPKT VVYKREEHYN DNISIKERNV SMVRKFGIEK HRLPQCHFLS | 1560 |
| NGNYDLMITN RGSGYSKRDN IFINRWREDI TDDNLGMFVY VKDIKSGVYW SNFYQPCKKE | 1620 |
| TKNYEVSFSV DKAKFERADG EIKTTTEICV SNEENGEIRK VTIDNNSGED KILEITSYFE | 1680 |
| VVLNKYDADL VHPSFSNLFI NTEFIEEPLC ILANRRKRSV EDKELWIMQS LITDENICGN | 1740 |
| VQYETSRANF IGRCRDLNRP EAIENDRIMS NTVGAVLDPI ISMRVRVKIP AKQSCQLSYV | 1800 |
| IAICENRQQC IEIADKYQQR DNVERVFQLA WTYSQIESRY LGIKSYMVNK FQELASNIIF | 1860 |
| PNESIKQREQ YVKNIRKNQV DLWKYGISGD IPIACIVIDK LEDIGCIKDI IKAHEYWNVK | 1920 |
| GLEVDLIIIN LEEYSYEQEL QISIKNIIFA SHLRYKEYTS GGVFLYNKST MLKEDIDFII | 1980 |
| SISRLVIYCE DERVLNEPKK NKKLIDKNKK INEIKNLKYN EGDFQFETPE LEYFNGFGGF | 2040 |
| KKASYEYIII LDNYKNTPAP WINVISNGKF GFHISENGVS YTWYKNSREN KLTPWSNDWV | 2100 |
| KDNQGEFIYL KDEETYDIWS ISPKPKRNSG KYIIEHGFGY STYRNKSFGI VGEMTSFVPL | 2160 |
| EDNVKVILVN LKNNTDIKRE ISVTYYAQLT LGVVSQHTSR YISTYKNDDY DYIYAKNPYN | 2220 |
| TYFGDLIAYC KVIGGEQISY TGDRMEFIGV DGSLENPNGL KNDMLSNTLG SGMDPCIAIN | 2280 |
| EKVILNPGEE KTVFILLGQE DNIEKIDSVI EKYENEESVL NELKRVKSYW EDTLTQIQIE | 2340 |
| TPDKSMDIMV NKWLLYQVIS CRFWARTGFY QSGGAYGFRD QLQDSISISY VRPELTREHI | 2400 |
| LYSASRQFLE GDVQHWWHPV VDSGIRTRFS DDLLWLPYVT IQYIKNTGDY EILNESINYL | 2460 |
| EDELLKEGED ERYTISKVSQ NKGSLYEHCI KAIDKSLKFG ENNIPLMGSG DWNDGMSTVG | 2520 |
| NKGKGESVWL GWFIYNILKE FTSICNKKQD YEKEEYYNKM KRFIVDNIEK NAWDGKWYRR | 2580 |
| AYFDDGKVLG SAENEECQID SLAQSWAVIS EGGNKSRAEK AMLSAEERLV KYDKAIVKLL | 2640 |
| TPAFNKSDLE PGYIKGYVPG VRENGGQYTH AAIWYILALA KLGYGEKAWK IFNMINPINH | 2700 |
| TLSNLNCNIY KVEPYVMTAD VYTVEPNVGR GGWSWYTGAA GWMYTTALNG ILGFNIHSDK | 2760 |
| GFSITPNIPS HWNEFKIVYK KNNCKYNIKV ERQGKEEVYL NNELIKDKII PLFNQGEYEV | 2820 |
| KIFIK | 2825 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1742 | 2818 | 0 | 0.9942084942084942 |
| GT84 | 1291 | 1503 | 1.8e-104 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1742 | 2825 | 1 | 1053 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.10e-162 | 1496 | 1829 | 1 | 336 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam17167 | Glyco_hydro_36 | 8.97e-156 | 2329 | 2753 | 1 | 423 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 9.32e-143 | 2030 | 2313 | 1 | 283 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 5.25e-97 | 2058 | 2298 | 1 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SNV88157.1 | 0.0 | 1 | 2825 | 1 | 2825 |
| AAO36920.1 | 0.0 | 1 | 2825 | 14 | 2838 |
| AVP54577.1 | 0.0 | 1 | 2825 | 14 | 2841 |
| CDI50658.1 | 0.0 | 633 | 2825 | 1 | 2193 |
| QBD85765.1 | 0.0 | 633 | 2825 | 1 | 2193 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 4.38e-87 | 2037 | 2817 | 3 | 802 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 4.43e-86 | 2037 | 2820 | 3 | 796 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 2.40e-66 | 2037 | 2794 | 3 | 787 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 3.38e-65 | 2037 | 2794 | 3 | 787 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 3AFJ_A | 3.68e-63 | 2037 | 2794 | 23 | 807 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 32 | 2807 | 67 | 2825 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| Q9F8X1 | 6.98e-91 | 2037 | 2805 | 3 | 781 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| B9K7M6 | 2.50e-86 | 2037 | 2817 | 3 | 804 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q76IQ9 | 1.58e-85 | 2037 | 2822 | 3 | 798 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 9.90e-54 | 2105 | 2811 | 84 | 771 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000068 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
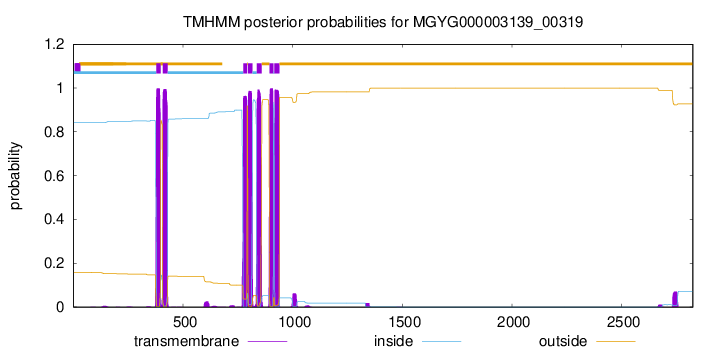
| start | end |
|---|---|
| 379 | 398 |
| 408 | 430 |
| 775 | 792 |
| 797 | 816 |
| 837 | 859 |
| 895 | 912 |
| 917 | 939 |
