You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003280_00653
You are here: Home > Sequence: MGYG000003280_00653
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-269 sp900762425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; CAG-269; CAG-269 sp900762425 | |||||||||||
| CAZyme ID | MGYG000003280_00653 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9712; End: 17889 Strand: + | |||||||||||
Full Sequence Download help
| MKILKINGTP LDKKQLEIHL QKVASNHILI SKSQKNTYPI PHLMESFKVI QNVYNILNEH | 60 |
| LKLGINIHPA GEWLLDNLYI IEESVKQIEK ELTVKKYQSF VGIANGPYEG FARIYVLASE | 120 |
| IVAYTDNKID RKDLEDYLLS YQTKKTLSME EIWNIGLFLE IAIIENIREV CERIYIAQME | 180 |
| KYKAENIVER LVENKTKSEQ IFKINTNKKM EKTIFSDMHY SFVEYMSYIL KRYGKKGYSY | 240 |
| LKALEETIEL TGTTVSDVIK KEHFDIAVRK VSIGNSITSI KKIQRINFLE IFEKINGVEE | 300 |
| LLKQDNSKIY EKMDSKTKEY YRNQIKEISK KTKISEIYIA KKLLELSNIH QKHIGYFLID | 360 |
| DGIEELYQTL NYKTNKKLNS KIKSKIYISI ITIFTLIISC YFAKIANYQI NKTCFFIFSM | 420 |
| IIFFIPASEI VIQIISYISS KIVKPKLIPK LDFSNGIDKE NKTMVVIPTI VKDKEKVNEL | 480 |
| MRKLEVFYLA NKSQNIYFCL LGDCSQSDKI EEKYDEEVIK EGKLLVENLN KKYKNENEMI | 540 |
| PIFNFIYRKR IWNDKENAYL GWERKRGMLN QFNEYLLGHI KNPFKENTIE NYIEKDNNLR | 600 |
| EELKNIKYII TLDSDTDLIL NSAFELVGAM AHVLNKPVLN EDETKVISGY GIMQPRIGIN | 660 |
| LNISFFNIFT KIFAGAGGID CYTNAISDIY QDNFQEGIFT GKGIYDLKIF SKVLNNKFPE | 720 |
| NTILSHDLLE GCYLRCGLVS DILLMDGYPT KYNSFMNRLS RWIRGDWQIT RWLLPKYGLN | 780 |
| LISRYKILDN LRRSLLEISV IIGAILLAVV KKSYLWVLII PLMPFILELV NLIIFRKEGE | 840 |
| QKSKTFTPKI AGVPGILLRA ILTIGCLPYK AYLSAKSIIK TLYRVIVSKN HLLEWMTSEE | 900 |
| AEKQAKSDNL SYIKQMFFNI LIGLIQIAYS IYSGNKYQSI LGILWIITPT IMCNISKRKN | 960 |
| KTKVVEKLSK EEKEYIKEIG RKTWGFFETY LNKENNYLIP DNYQEDRKEK IVNRTSSTNI | 1020 |
| GLSLLAVISA YDLKYISKEK SIELIKNIIY TISDLPKWNG HLYNWYQIKT KEPLIPRYIS | 1080 |
| TVDSGNFVGY LYVVKSFLNN ILKEYRESQI NIENNKNESN NKITDNNFIE IENLIDIIES | 1140 |
| LIKNTDFSVL YNNELQIFSI GFNIEENKLT DSYYDLLASE SRQASLVAIA KKDVPAKHWN | 1200 |
| YLSRTLTTLG KYKGLISWSG TAFEYLMPDV NIPRYEGSLL DESCKFMIMS QKEYAKKLNI | 1260 |
| PWGISESAFN VKDLKANYQY KAFGIPWLGL KRGLADELVV SSYGGILAIN DEPKEELENL | 1320 |
| KILEKEGMYD KYGFYESIDY TPERVDKGKT SSVVKTYMAH HQGLILLAIN NFFNNQILQK | 1380 |
| RFMENPDIEA VSILLQETMP ETSIITKEKK EKVEKLKYKD YENYIKTDYN KIDERLITGN | 1440 |
| VISNENYTVA INQKGEGISK YKNIYVNRFK KTDDYSQGIF FVMKNIKTKQ IWSSNYKFNN | 1500 |
| ENDKKYQISF MPDKVEQEII EGNIKSKIRT TVASNEPVEL RRATIENYGN EEEIIEVTTY | 1560 |
| FEPVLSIKEQ DYAHPVFNNL FLMYEYDEET NSIVIKRKNR EENSKEIYLM ANLSTNSETI | 1620 |
| GDLEYEINKS RFIGRGNLGI PSMVQNSIPL SKKIGLVTEP IVALKRIVKI KPGQKIDVDL | 1680 |
| VISVGEEKQK VKENIEKYQS LKNIKMEFEL SKARIEAESR YLRIKGKEIA LYQKILSYII | 1740 |
| FDNSVKSQRV KNIKQNQYKQ SDLWKYGISG DLPVILVKMR DVNDSYVIKE VLKAYEFFRT | 1800 |
| KNVQTELIIL DEEKHSYENY VKEEIENAIL NQHIAYLKNQ KGGIFSLVKS EMDEQDINLL | 1860 |
| EILATIVIDS SKGTLENNIK DIEEEYFEKN KILEEEHQLL LEKENTNDID ILENNKELKY | 1920 |
| YNEYGAFSPD GKEYLIKVNK QNRLPTVWSH ILTNQNFGTV VTENLGGYTW YKNSRLNRVS | 1980 |
| SWQNNPSYDI PSEIIYLKDM ETKKTWSLGL NPMPDNNNYN IIYGFGYCKY IHKSDGIEQE | 2040 |
| LEIFVPTEDS CKVGILNLKN ITPNRKKIKL YYYIKPVLGE DEIKTNGYIC TNFDKNNNII | 2100 |
| YSKSIYNTED KSSIAYISSS EKIISYTGSK KSFLGSGKIE NPIGIKKEKL NNETGLGQES | 2160 |
| CMALEINIEL ESYANKEVII MLGANDNLID CKNTAYKYSK LSNVKQELLK TKNYWKEILG | 2220 |
| RLQVYTPLES TNILLNGWIP YQTIASRLYA RSGYYQSGGA YGFRDQLQDT LGVRYLEPEF | 2280 |
| MKNQIIKHSK HQFLEGDVEH WWHEETGRGI RTRFSDDLLW LVYLTIKYIE FTGDNSILDI | 2340 |
| ETKYLSGEIL QEGQDEKYDL FKQSDVSESI YEHCIRAIEK SLNFGEHGLP KIGSGDWNDG | 2400 |
| FSTVGNKGNG ESIWLGFFLY TILDGFIPII KERENDGNEN ENVIEEKIKL SEKYEKIKQD | 2460 |
| LKRALNTSGW DGRWYKRAYM DDGNILGSME NDECRIDSIA QSWSVISNAG ENDKKYISME | 2520 |
| SLENHLVDRE NGIIKLLDPP FEKGKLQPGY IKAYLPGVRE NGGQYTHAAI WAIIAESILG | 2580 |
| FGDKALELYR MINPIEHART KDACNKYKVE PYIIPADVYG AGNLIGRGGW TWYTGSSSWY | 2640 |
| YEAGIYYILG LVIEDGYITI KPCIPKEWKE YKIQYKWKES IYNIVVKNPD AKNREVRKIM | 2700 |
| LDGKEIENKI KLDGERKVYN IEVIM | 2725 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1624 | 2722 | 0 | 0.9980694980694981 |
| GT84 | 1172 | 1386 | 7.3e-91 | 0.9953488372093023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 0.0 | 1624 | 2724 | 1 | 1051 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 3.41e-126 | 2214 | 2651 | 1 | 424 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 1.89e-86 | 1917 | 2187 | 1 | 272 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 1.44e-78 | 1945 | 2183 | 1 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.98e-65 | 1378 | 1710 | 1 | 336 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUG57227.1 | 0.0 | 3 | 2725 | 31 | 2909 |
| AEV69563.1 | 0.0 | 4 | 2725 | 32 | 2919 |
| ADU74104.1 | 0.0 | 4 | 2725 | 36 | 2919 |
| ANV75789.1 | 0.0 | 4 | 2725 | 36 | 2919 |
| ABN52453.1 | 0.0 | 4 | 2725 | 36 | 2919 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 2.55e-92 | 1924 | 2725 | 3 | 810 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 1V7V_A | 4.50e-82 | 1924 | 2710 | 3 | 785 | Crystalstructure of Vibrio proteolyticus chitobiose phosphorylase [Vibrio proteolyticus],1V7W_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc [Vibrio proteolyticus],1V7X_A Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate [Vibrio proteolyticus] |
| 3S4C_A | 1.58e-61 | 1924 | 2710 | 3 | 806 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 2.20e-60 | 1924 | 2710 | 3 | 806 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 3AFJ_A | 2.29e-57 | 1924 | 2691 | 23 | 808 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 0.0 | 45 | 2695 | 64 | 2813 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 5.36e-90 | 1924 | 2725 | 3 | 812 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q9F8X1 | 9.81e-84 | 1924 | 2707 | 3 | 782 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q76IQ9 | 2.19e-81 | 1924 | 2710 | 3 | 785 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q7S0S2 | 2.56e-38 | 1991 | 2697 | 83 | 764 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
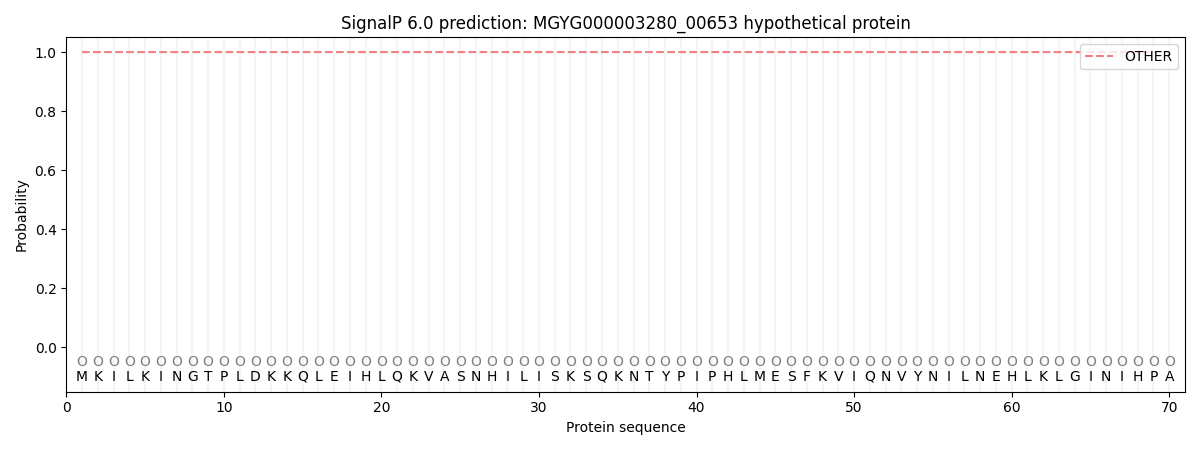
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000033 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
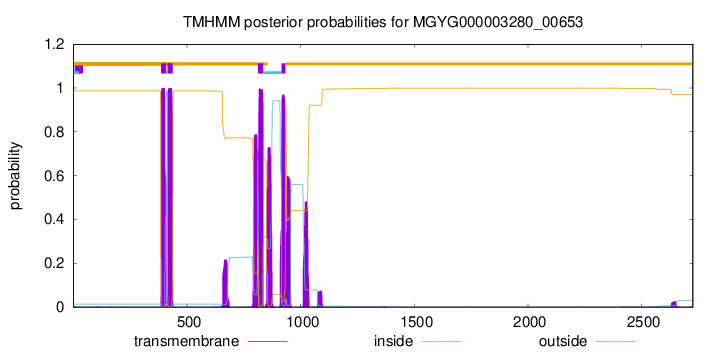
| start | end |
|---|---|
| 386 | 408 |
| 415 | 437 |
| 813 | 835 |
| 916 | 933 |
