You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003987_00597
You are here: Home > Sequence: MGYG000003987_00597
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1691 sp900544375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UBA1691; UBA1691 sp900544375 | |||||||||||
| CAZyme ID | MGYG000003987_00597 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10533; End: 17879 Strand: - | |||||||||||
Full Sequence Download help
| MKRDIIRRAK PRALKRQIEK DLYSLRKKYT KALRAHDDTA LGEWLCDNFY LLEREGRGVL | 60 |
| SQLKTAPALP LDADGVVRIY ALCLRAVREH ADLSEQILES GLQQAGLCSA EVELVPLFLR | 120 |
| AALLHTARLG CEETGEPAVQ RLSHAVLTLR AMQDWDFDML LESLSPVEQI LMRDPEGIYP | 180 |
| QMDETTRFCY RRLTAQAARN AGITEQEAAC AALSQAQAGE TPGERHVGAW LPSGPSYRRR | 240 |
| GKTALLMEAL FPLLVALCTA ILTRAWYLVP LLYFPVWEIL RYPIEAAFSS GVAVRPMHRL | 300 |
| NLEGMIPEEG ATLIAVSTLL PGASQARKLG QHLEQLWRTN GHGSVKICVL ADLKGAQSPT | 360 |
| MPQDTSDLAA SRREIEQLNR RHNGGFVLIV RERSYSRTQE EYTGAERKRG AIEALIRYLR | 420 |
| GEEVPFRALC GDTDGLQNTK YVLALDADTV LPLDAASQLV AAAMHPLNRP VVDEKKGIVT | 480 |
| QGYGILVPRV ETELFSAGAT GFSRVMADAG GLVAYDTAAA ERYQDFFGRS IFSGKGLIDV | 540 |
| DVFYRVLLGV FEPESVLSHD ILEGGFLRAG FVSDVQVADG FPGREGAYLD RMHRWVRGDW | 600 |
| QNLPFLIRRK NRKGGQKGSP LGALARWQLF DNLRRSFTPV SALLCLIAAL FTPAPARLLL | 660 |
| VLGGLLCTMA GSLYGAWRAV LSGGPGMFSR LYYSRVTPVA TGNLIRAIVS VVMLAQTAFV | 720 |
| SLDAIIRALW RRFVSHKKLL EWVTAAQSDV AQRPGALVKR YLPSILTGAL LMLFGRGSLW | 780 |
| LCGFLFLCNF PFAVFSARSG VEKTKELNWE QRDRLTSYAA AAWRYYEEFC GVQDHYLPPD | 840 |
| NVQETPVHRV AHRTSPTNIG LALLCTLAAR DFSFIDSAML CERIDGMLTS IEKLEKWHGN | 900 |
| LYNWYDTTTL RVMEPRYVST VDSGNFFCCL TALRQGLLEY AAEEPKLVDQ ANRVGRLLEE | 960 |
| GDLSPLYNPR RRLFHIGFDA QEKKLTGSYY DLLMSEARMM SYYAVGSRQA PKKHWGALGR | 1020 |
| MLAREGRYTG PVSWTGTMFE YYMPHLLLPL YDGTLGKEAM RFCSYCQKKR VRGKNIPWGI | 1080 |
| SESGFYAFDP QLNYQYKAHG VQKLGLKRGL NDDLVISPYS TFLLLPFEPE AALKNFDELE | 1140 |
| QMQMTGRCGF YEAADFSPER VDGQEYALVR SYMAHHVGMS MVSVCNALKG YAMQNRFMRD | 1200 |
| DRMAAARCLL EEKIPTGASV FHDVELRETP QRAQRVTSAT REIMEPNPVQ PQMHLLTNGE | 1260 |
| WSVAVSDCGT SVSLYRESDI TWRGGDLLRR PKGIFAVARA QEETLPLCRA LDYRSGAEFS | 1320 |
| AQFSHTQARL TAWSKTLVGE TLIQVHPRLP CEHRRYTVKN LGKERAHISL LIYLEPCLAP | 1380 |
| AREAKAHPAF SKLFLEHGYD AGNKLQLFTR RPRGDGEPMC LAAGLLEDVP FHYEGAREKL | 1440 |
| LDFPDGIFTL RNAPMKLGDG IGVGDPACAF SVSLEIEPRA ERHVTLLLAA ASTRAEAADR | 1500 |
| LLRSRREGRI VAGAPNPFYE NSLDGILAAQ VLPQIFYPPQ ETNEYLAAAQ ENNAGVQALW | 1560 |
| SLGISGDHPI LYIQIQNAED VARALPLVRL NRKLRRSGIP TDVAIGYREG GAYDRPVLKA | 1620 |
| LETALRHENC IESLNAPGGI HAVDLQTHTA QSVLALQTAA RYISPSAAER MQLPVLPFVP | 1680 |
| FAVFPVLPAQ RRLELDFCVR RGGFMENGAF AITEKPEVPW CHVLANLDFG TLVESGALGC | 1740 |
| TWAVNARENK LTPWFNDTRT GNRGELLFLR VKNEVYDLIQ GARAEFSPGM ALWRGRAGKI | 1800 |
| ETAVTVAVPD TGMLKTCEVE FYNNGKEPVE VETVYYTEPV LGVDRQNARF IKSRWQGGVL | 1860 |
| SLHSPWNTAV PGYMALTGGE GDGAFCCCDR ADFWRGKWGE QRMLPLDDPC AAVGRRLVLP | 1920 |
| PKRKEKVRFV LSFAAQEQAA KALLEIAPKR VLHNALRVDT PDKALNHMVS SFLPAQILNS | 1980 |
| RIFGRTAFYQ NGGAWGFRDQ LQDVGSMIFL RPRLARQQIL RAAACQFPEG DVLHWWHRLP | 2040 |
| GKNGLRGVRT RYSDDLLWLP YVTAEYIKQT GDASLLHLKI PYLEGEALRD GEHERYFEPT | 2100 |
| MSAQRGTLYE HCLRAIEHAA RFGAHGLPLI GGGDWNDGFN RVGIQGRGES VWLAQFMALT | 2160 |
| LDEMRPVCKL MEDESSEARF AQRAQELRSA VDETAWDGRW YRRAYFDDGT PMGASGAPAC | 2220 |
| EIDSLPQSFS VLGGMPDEQR RNLALDSAVE KLVDWEQGLI RLFTPSFTGE GYSPGYVAAY | 2280 |
| PAGVRENGGQ YTHAAVWLCM ALLREGRVDE GYSLLKLLNP AEKCAQEAQA KRYLREPYAL | 2340 |
| AGDVSTAKGI EGRGGWSLYT GAAGWYYRTV VEELLGIRLR SGALELEPRL PSGWHGYSAT | 2400 |
| LTLDGAVIAI TVSDTAPQGL TVDGEQAERF TPDGREHRLV YGIGASAS | 2448 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1528 | 2438 | 3.1e-232 | 0.890926640926641 |
| GT84 | 988 | 1200 | 1.8e-79 | 0.9906976744186047 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 6.85e-173 | 1558 | 2440 | 131 | 1050 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 2.39e-102 | 1954 | 2378 | 7 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 3.55e-37 | 1694 | 1945 | 1 | 281 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 3.74e-37 | 1193 | 1485 | 1 | 305 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 1.98e-26 | 1718 | 1931 | 2 | 246 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI59942.1 | 0.0 | 13 | 2410 | 31 | 2442 |
| AVQ95160.1 | 0.0 | 9 | 2443 | 36 | 2551 |
| AYF37850.1 | 0.0 | 9 | 2443 | 36 | 2551 |
| QCN91406.1 | 0.0 | 9 | 2443 | 36 | 2551 |
| AYF40573.1 | 0.0 | 9 | 2443 | 36 | 2551 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QDE_A | 1.46e-48 | 1923 | 2439 | 281 | 806 | ChainA, Cellobiose phosphorylase [Acetivibrio thermocellus],3QDE_B Chain B, Cellobiose phosphorylase [Acetivibrio thermocellus] |
| 3AFJ_A | 3.37e-47 | 1933 | 2431 | 318 | 826 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
| 4ZLE_A | 5.26e-47 | 1956 | 2411 | 312 | 745 | Cellobionicacid phosphorylase - ligand free structure [Saccharophagus degradans 2-40],4ZLF_A Cellobionic acid phosphorylase - cellobionic acid complex [Saccharophagus degradans 2-40],4ZLG_A Cellobionic acid phosphorylase - gluconic acid complex [Saccharophagus degradans 2-40],4ZLI_A Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex [Saccharophagus degradans 2-40] |
| 2CQS_A | 5.96e-47 | 1933 | 2431 | 318 | 826 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3ACT_A | 4.36e-46 | 1933 | 2431 | 318 | 826 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127],3ACT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 2.57e-317 | 142 | 2419 | 285 | 2812 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 1.96e-50 | 1923 | 2439 | 282 | 808 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q7S0S2 | 1.82e-40 | 1907 | 2412 | 255 | 753 | Cellobionic acid phosphorylase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU09425 PE=1 SV=1 |
| Q9F8X1 | 2.24e-37 | 1706 | 2444 | 9 | 800 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
| Q76IQ9 | 7.58e-34 | 1957 | 2444 | 305 | 801 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000058 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
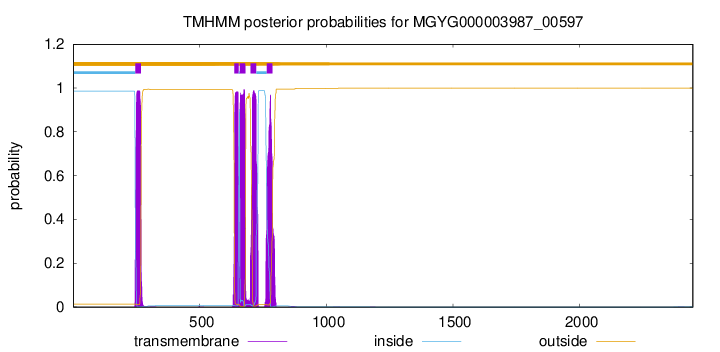
| start | end |
|---|---|
| 245 | 267 |
| 636 | 653 |
| 658 | 680 |
| 700 | 722 |
| 765 | 787 |
