You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004074_00344
You are here: Home > Sequence: MGYG000004074_00344
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
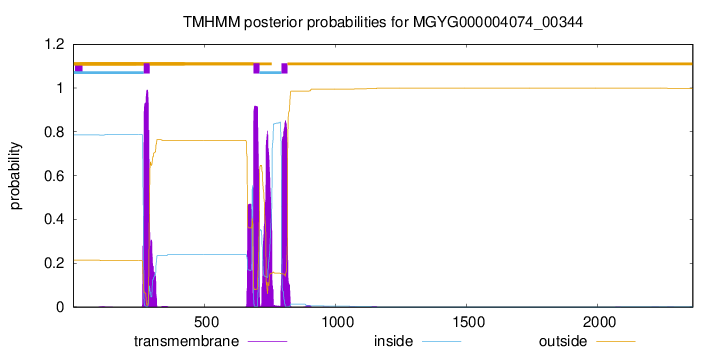
TMHMM annotations
Basic Information help
| Species | HGM12998 sp900756495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; HGM12998; HGM12998 sp900756495 | |||||||||||
| CAZyme ID | MGYG000004074_00344 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20999; End: 28093 Strand: - | |||||||||||
Full Sequence Download help
| MAEQWERLGE DLARNLVPAG QTSGRRAAGE IRAALRKLDR LRRRQAGQEE QTQAAEWLLD | 60 |
| NWYVAQREGL EGMRALRSAR RLRFVQQAGG RELYVLELCR ALAGTAEPLD AAVIAAFLAG | 120 |
| VQSVRPLTEG ELAHFFPALR GALVLQLLKT AECLDRGEKV PAEVMEQIFA SLRALSAPGL | 180 |
| WKLLEQASIV DQVLRRDPAG AYPGMEDATR AQYRSTLCML ARRYGMSEAA AADLTLRLAQ | 240 |
| SGDGERRHVG YYLYRRPLGR QNRQNGGTLY VAGILLPTLF LVLLIGFHLH SWLVTVLLLL | 300 |
| PVSDIIKNLM DFLVVRLVPP RPVSRMALEG GIPEDGRTLC VIAGLLTDNQ GGPVYAALLE | 360 |
| RYRLANRDAG KHLLMGLLAD LPDRDTPVDG RARGWIREAN RAVAALNEKY GGGFYLFFRE | 420 |
| PVFQPGEERY QGWERKRGAL LELCRLLRGK RTGVRVEEGD RRCLRGVKLV ITLDSDTSLN | 480 |
| VGTARELAGA MLHPLNRPTV DPRRRIVTAG YGVLQPRVGV ELGSANRSWF SRIFAGQGGV | 540 |
| DPYGGTCSDV YHDLFDQGTY TGKGIFDVDA FLTCLDGRFP ENRILSHDLI EGSYLHCGLL | 600 |
| GDIELTDGYP YKVTSYFARL HRWVRGDWQL LPWLSGTVKN QAGRREANPI SSVAKWKIFD | 660 |
| NLRRSLSPVC TLLALILGMC LHRTDFGWAA GIAVIAAASH LLLSGAELAW RGGKGLRERY | 720 |
| HSTIVSGLGG AVLQTLLQLL FLPLHAWICF SAICTSLWRQ LVSHRNLLAW VTASAAEQRA | 780 |
| GNGLWANVRK QWPAVVIGLA AMLFSRFPAG AAAGLLWMLS PVFSWALSRP IREGRRVADA | 840 |
| DRPFLLHQGA LIWRYFEDFL RPEDHWLPPD NWQEQPPVGL ARRTSPTNIG MALLSVMAAA | 900 |
| DLEYISQEKA INLLSHTIST LEILPKWRGH LYNWYDTSEL SPLYPRYVST VDSGNLCACL | 960 |
| VALREGLYQW GAAALARRAE ALSDAMDFSG LYDRERRLFY IGYDLEKEEY TQGWYDLLAS | 1020 |
| EARQTSYLAV ARGEVEPRHW RRLSRTLTAD GDYRGMASWT GTMFEYFMPN LLMPLETNSL | 1080 |
| LYESLCFCVW EQRRRTAGKY PWGISESCFY AFDPGMSYQY KAHGVQRLGL KRGLDCELVI | 1140 |
| APYASFLSLP AAPSAAVKNL RRLRELGAEG MYGLCEALDF TPSRLTGDRP FEPVRTYMSH | 1200 |
| HLGMSLVAID NALRENVMVR RFMADCNMMA YRELLQEKVP VGAVVLKSQT GDAPERPRRI | 1260 |
| TEEGVVREGI WTGEGRPACH LLSNGQINCF CAADGSNRTV WREFLLTGGD GISCYLYAEG | 1320 |
| AVSPLTPNSW RFDGGASWTT ETEGFTALRI LRTAEKAPGV SWEVRLRRKG NEPLPGELVL | 1380 |
| YLEPILARKR DFEAHPAYVK LFVESAYTGD GIVFARRPRR QGEPVPALAV VWDAETAFFD | 1440 |
| TSRESALGRG GLSALPQALR RQARSSAGAV LDPCLLLRIP LTLSPGRTSA FRFALAISYS | 1500 |
| GEEALEEARA LLHEGTAGTD KNQNRLIELC GLTGETALQA FELLKVLSYP GELPNTPQSD | 1560 |
| LWPYGVSGDY PIALIRAASR EEAEKHAFWL GCHKFLTHLG YPFDLVILLE EGGDYRRPAR | 1620 |
| SLLFETLREL HWDHMQGARA GIHLLQGPAP ALEAAAIAAD GPYPPFKNTA HTGISSFLLD | 1680 |
| SGSPLWETKP DGTFTLHCGP RLPPLGWSQM LTNGGLGWMP DETGCGHLWL GNARECQLTP | 1740 |
| WCNDPLQIGG PEYLSLLVGE EERSLFAAAD GLPCDVTYGP GFARWTKKWG EHRTVLTACI | 1800 |
| PMEQDVRILL LEMEGTPAQV KYRHPEGTEH LFPIREAIAI LTDSGGVTRT GDVREAKKML | 1860 |
| NRTVMHWNRT VSALKFHTPD EALDNYLNGW ALYQVMACRL MGRTSRYQNG GAYGFRDQLQ | 1920 |
| DACAVLFSDG TIARTQIRKA CAHQFEEGDV LHWWHETPGA ADKGVRTRIS DDLLWLPYAL | 1980 |
| CEYLEKLGDD GLADEIVPYL TSRPLEAEEQ ERYEHAEISR ETGRVYDHAC RAADLVLERG | 2040 |
| TGEHRLLLMG AGDWNDGMNR VGAAGKGESV WLSWFAAHVL GRLAALCAAR GDGSRAESYR | 2100 |
| AAAEKLRSAA GAAWDGDWFL RGYYDDATPL GGRQCDECRI DSIAQSWAVL AGSGEERAQI | 2160 |
| AVRSALEALW DQSAGLVRLF TPAFDDGEKD PGYIKGYLPG VRENGGQYTH AAVWLALACL | 2220 |
| ELGMTSEGYG ILKNLLPAHH DPEIYKAEPY VLAADVYDNP AHRGRGGWSW YTGAAGWYYR | 2280 |
| TATEYLLGLK LRAGRLSVEP KLPPDWPGFT AVWRTEQATL RISVSRGKEK KSLLDGEPVQ | 2340 |
| EGIPIRDLSG EHKLEVVIQA SGSI | 2364 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1851 | 2356 | 1.1e-185 | 0.4806949806949807 |
| GT84 | 1014 | 1225 | 3.8e-80 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 5.01e-164 | 1439 | 2358 | 2 | 1052 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 3.06e-108 | 1866 | 2290 | 1 | 425 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.85e-30 | 1218 | 1506 | 1 | 319 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 2.22e-20 | 1362 | 1488 | 110 | 239 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| pfam10091 | Glycoamylase | 3.75e-13 | 1013 | 1224 | 1 | 213 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBB66089.1 | 0.0 | 7 | 2354 | 16 | 2395 |
| ALP93881.1 | 0.0 | 7 | 2040 | 16 | 2084 |
| CDZ23197.1 | 0.0 | 10 | 2345 | 15 | 2486 |
| AVQ95160.1 | 0.0 | 54 | 2345 | 69 | 2540 |
| AYF37850.1 | 0.0 | 54 | 2345 | 69 | 2540 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZLE_A | 1.60e-48 | 1874 | 2361 | 312 | 787 | Cellobionicacid phosphorylase - ligand free structure [Saccharophagus degradans 2-40],4ZLF_A Cellobionic acid phosphorylase - cellobionic acid complex [Saccharophagus degradans 2-40],4ZLG_A Cellobionic acid phosphorylase - gluconic acid complex [Saccharophagus degradans 2-40],4ZLI_A Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex [Saccharophagus degradans 2-40] |
| 3S4C_A | 3.53e-46 | 1866 | 2340 | 314 | 803 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
| 3RRS_A | 1.95e-45 | 1866 | 2340 | 314 | 803 | Crystalstructure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RRS_B Crystal structure analysis of cellobiose phosphorylase from Cellulomonas uda [Cellulomonas uda],3RSY_A Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3RSY_B Cellobiose phosphorylase from Cellulomonas uda in complex with sulfate and glycerol [Cellulomonas uda],3S4A_A Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4A_B Cellobiose phosphorylase from Cellulomonas uda in complex with cellobiose [Cellulomonas uda],3S4B_A Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda],3S4B_B Cellobiose phosphorylase from Cellulomonas uda in complex with glucose [Cellulomonas uda] |
| 2CQS_A | 7.08e-45 | 1866 | 2358 | 334 | 842 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3AFJ_A | 9.40e-45 | 1866 | 2358 | 334 | 842 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 3.68e-317 | 165 | 2339 | 285 | 2815 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 3.26e-47 | 1801 | 2358 | 253 | 812 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
| Q76IQ9 | 8.91e-45 | 1866 | 2358 | 296 | 800 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio proteolyticus OX=671 GN=chbP PE=1 SV=1 |
| Q9F8X1 | 1.92e-41 | 1866 | 2358 | 296 | 799 | N,N'-diacetylchitobiose phosphorylase OS=Vibrio furnissii OX=29494 GN=chbP PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
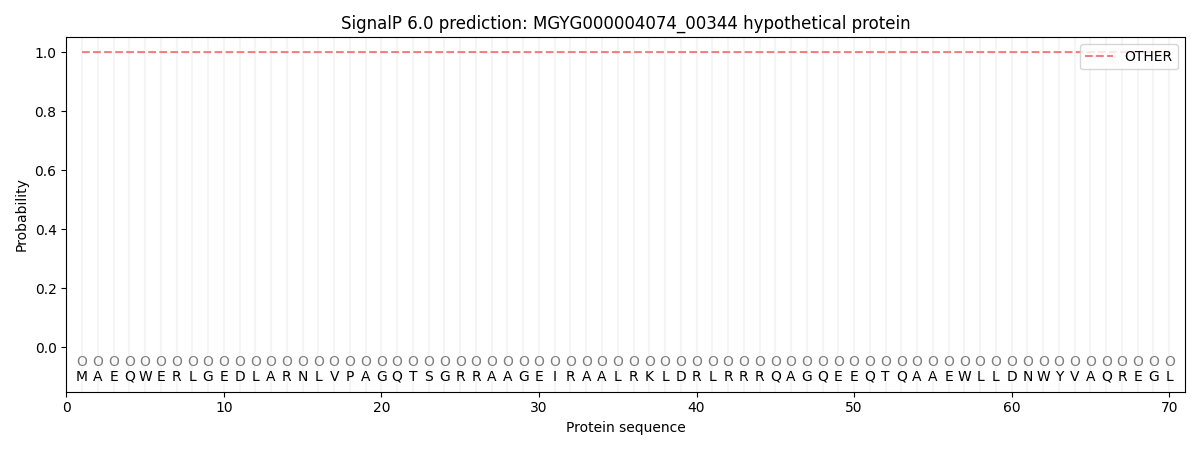
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000042 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |