You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004526_00470
You are here: Home > Sequence: MGYG000004526_00470
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
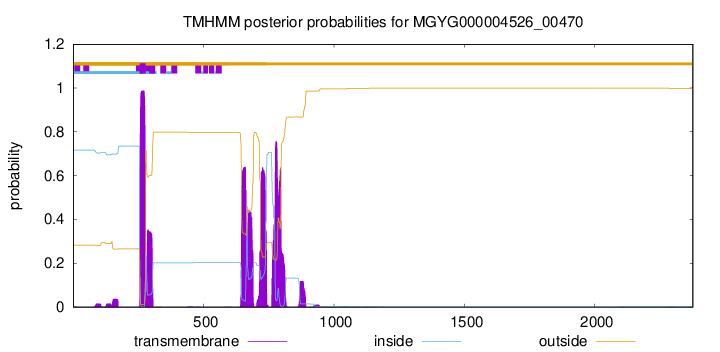
TMHMM annotations
Basic Information help
| Species | CAG-103 sp000432375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-103; CAG-103 sp000432375 | |||||||||||
| CAZyme ID | MGYG000004526_00470 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42873; End: 50003 Strand: + | |||||||||||
Full Sequence Download help
| MAETNTAALR AQGERDARAL TVTGRRRADT GRIRRAAKTL ARMARAVTPE TPNGQWLRDN | 60 |
| RSFACAAAGD AVAALRHARV RASGGQTALG ACCAGLLRAC GGALTVKAAE AYLEGFQDAL | 120 |
| PLETAELALL VPGLQAAVVC ALAESYAGDG AAAPALFTSL RALGTAAWGM LAERCDRVGR | 180 |
| ILARDPVGVY PAMDAATRAH YRQTVARLAR RTGRTEIEIA EDVLARAQRS EGARRHVGWF | 240 |
| LLREVLGAPE RRRDGGWYIA AVVVGTLFLS LLLGFATKSV SGAVLLLIPV SEVVKGLLDA | 300 |
| VLLRVTKPRF VPRLALRRGI PPEGRTLCVI AALLTSPDDA HALCARLEEY YLCNRDALGQ | 360 |
| LAFGLLADLP EADIAHAPID PAILRAAREE IAALNARWGS RFYLFTRPRV QTPDGKWSAW | 420 |
| ERKRGALLEL ARLVLDRPGT LQCAAGDAAG LSGTVYLLTL DADTRLTPGA ARALVGAMLH | 480 |
| PLNRAVVDAQ RGVVTRGYGL LHPRMATELQ SANATDWARV FAGPGGADPY GGACGELYMD | 540 |
| CFDRGGFPGK GILDVRALLD CCGGGVIPEG RVLSHDALEG AYLHGGFLGD VELTDTFPAA | 600 |
| PLAWGARAHR WIRGDWQNAP WIFSRRARAL HPIDRFRLAD NLRRSLVAPA TWISIFLGCV | 660 |
| LRWPGLRLAA YAALLALAQG LIRALGQPIV HPADTRVRYH SDVLHGLASA VAQTVLRLIL | 720 |
| LPWEAILNTS AIVTALWRMA VSHRNLLQWQ TAAQRAGRRD GLGAYWRALW PASALGLLTA | 780 |
| VLTPSPTGLA AGIVWMLSPF VLAALGAPNA AGAAPLSRAA QRELLGWAKA TWQYFETFCT | 840 |
| AEDHYLPPDN VQSQPPTGTA HRTSPTNMGL ALVSALGAHA LGIDDGQGLA LAERMLTTME | 900 |
| QLPRWNGHFY NWYHTCTLRP MPPLYVSTVD SGNCAAALLT AANALRGWGR DELAARAQML | 960 |
| CDGMDFGPLY DPQRRLMHIG IDTGSGKWSE SYYDLLSSEA RLTSYFAVAR GDVPREHWRA | 1020 |
| LSRAQVQKDH YRGCVSWSGS MFEYLMPELF LPPVRDSLLW ESAKFCLYVQ RRRVRPGQAW | 1080 |
| GVSESAYFAL DSALSYRYKA HGCAALALQP GMDKELVLSP YSSFLALAVE PRAAMRNLRR | 1140 |
| LAALGLLGQH GFFDALDCTR ARTGGGGQIV RCVMAHHQGM SLLAACNALC GDQVRRWFFA | 1200 |
| DPAMRAHATL LSERLPLGAP ALRRRAEQPA EKARRRALPD YAAEGTGIDA EAPRCALLSN | 1260 |
| GTYCLAVSEA GQSFARWHAL SVCRRGTRAD PAAGPVVRLE TDDFSGPLLP GTAAGAQWRF | 1320 |
| TVRAAQFTAR HGDVTAVQTL TVPGRANGEV RALTLSSPRG IEGTVRFVFE AVLAPLEDYV | 1380 |
| NHPAFARLGF QSVLRRDALL LRHLGRGEKP EAWLCAACDR PGTWRAEGPV PGWLRAGRVE | 1440 |
| LSVPVHTAPD TPWRVRFALG VGAREDDAFT AAQRALVLPP AMAAALPGEL AAQYGMTGAE | 1500 |
| LDGAMAAVSP LLFPAVAANA VPEQLERPAL WAQGISGDLP VWCAEAERGV VRQWALLRAL | 1560 |
| GVACDLALRS GDGGDYRQPT AGKVRGWLAA LDLAHTLGAP GGVHLVPEEA FPAVAAAAAI | 1620 |
| RPGMARTRCT LPLLPPPGDR RYLPTGSDVH WDDDGTVSFT GLPPRAWSQV LTNGRFGFLA | 1680 |
| TDAGTGHMWH RNAHTGRINR WLCDPWVLRG TETLRMASRA GAVSLFDDDG RSRVEYGFGW | 1740 |
| AAWERSVDGM SVRVTAFVPE DADARVLLIE CAGRARITWH TDLVCAGRDA DAPAVVTAYA | 1800 |
| DGLFTAENVR ADAPTLFCAA AGMPLTGWTC DRFSFLRGQM DARAGAGLSP CFALEGVIDR | 1860 |
| QGVLVCGCDT RANLLRLTQP DEAAHALRAT RERWLGAVSR LWMTTPDADM NRYLGGWAAY | 1920 |
| QTLCCRLLAR TSLYQNGGAF GFRDQLQDAV NLLLLDSAPA REQIGRACAH QFAAGDVCHW | 1980 |
| WHAGTGPEHG VRTRISDDLL WLPWAVCEYV EKTGDTEILS AEHPFLAGEE LEENEHDRYQ | 2040 |
| PLEVSAETGT VLEHCRRALM RVLARGVGAH GLLRIGTGDW NDAFDRVGAQ GRGESVWLTW | 2100 |
| FFAITARKFA ALVGGHAAEQ LALAADHCTR AAEGAWDGAW YRRGYYDDGR PLGSEACGEC | 2160 |
| RIDAIAQAFA ALDTHADPGR VHTALTSAVE HLFDREHNVV RLFDPPITRR TPETGYVRSY | 2220 |
| GAGLRENGGQ YTHGALWLAM ALLRTGRTAE GAEILHAVLP AAHDPARYEG EPYVLAADIA | 2280 |
| GGDNAGVAGW TWYTGAAGWY LRIAAEDLLG LHLRGGVLYP EPHLPAGWPE CTVRWRDGAG | 2340 |
| LLHTVRLRPD GVTVDNAPYD GGGIGKKRPK PYSQDR | 2376 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH94 | 1526 | 2361 | 9.1e-176 | 0.8474903474903475 |
| GT84 | 991 | 1202 | 1.5e-72 | 0.9953488372093023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3459 | COG3459 | 5.39e-137 | 1664 | 2352 | 293 | 1023 | Cellobiose phosphorylase [Carbohydrate transport and metabolism]. |
| pfam17167 | Glyco_hydro_36 | 2.03e-78 | 1894 | 2311 | 2 | 424 | Glycosyl hydrolase 36 superfamily, catalytic domain. This is the catalytic region of the superfamily of enzymes referred to as GH36. UniProtKB:Q76IQ9 is a chitobiose phosphorylase that catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. The full-length enzyme comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain. The alpha-helical barrel component of the domain, this family, is the catalytic region. |
| cd11753 | GH94N_ChvB_NdvB_2_like | 8.13e-22 | 1195 | 1388 | 2 | 205 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| cd11756 | GH94N_ChvB_NdvB_1_like | 1.73e-20 | 1663 | 1859 | 28 | 254 | First GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This first of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 8.35e-20 | 1664 | 1859 | 1 | 233 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBB66089.1 | 0.0 | 13 | 2362 | 18 | 2383 |
| ALP93881.1 | 0.0 | 13 | 2080 | 18 | 2102 |
| AVQ95160.1 | 0.0 | 99 | 2361 | 117 | 2542 |
| AYF37850.1 | 0.0 | 99 | 2361 | 117 | 2542 |
| QCN91406.1 | 0.0 | 99 | 2361 | 117 | 2542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2CQS_A | 6.10e-37 | 1872 | 2361 | 313 | 824 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate [Cellulomonas gilvus],2CQT_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],2CQT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate [Cellulomonas gilvus],3QFY_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFY_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and Isofagomine [Cellulomonas gilvus],3QFZ_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QFZ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Sulfate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_A Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus],3QG0_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Complexed with Phosphate and 1-Deoxynojirimycin [Cellulomonas gilvus] |
| 3AFJ_A | 2.46e-36 | 1872 | 2361 | 313 | 824 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127],3AFJ_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase triple mutant [Cellulomonas gilvus ATCC 13127] |
| 3ACT_A | 4.29e-36 | 1872 | 2361 | 313 | 824 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127],3ACT_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Histidine mutant [Cellulomonas gilvus ATCC 13127] |
| 3ACS_A | 9.87e-36 | 1872 | 2361 | 313 | 824 | CrystalStructure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127],3ACS_B Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase W488F mutant [Cellulomonas gilvus ATCC 13127] |
| 3S4C_A | 2.36e-33 | 1872 | 2361 | 293 | 804 | Lactosephosphorylase in complex with sulfate [Cellulomonas uda],3S4D_A Lactose phosphorylase in a ternary complex with cellobiose and sulfate [Cellulomonas uda] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 4.05e-286 | 156 | 2350 | 288 | 2808 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
| B9K7M6 | 3.43e-39 | 1876 | 2364 | 291 | 782 | Cellobiose phosphorylase OS=Thermotoga neapolitana (strain ATCC 49049 / DSM 4359 / NBRC 107923 / NS-E) OX=309803 GN=cbpA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000056 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |