You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002406_00130
You are here: Home > Sequence: MGYG000002406_00130
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium pacaense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium pacaense | |||||||||||
| CAZyme ID | MGYG000002406_00130 | |||||||||||
| CAZy Family | GT89 | |||||||||||
| CAZyme Description | Terminal beta-(1->2)-arabinofuranosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97779; End: 99734 Strand: - | |||||||||||
Full Sequence Download help
| MTLTAPRPDS ELVQERPKAP GITAVTAVLA ALTAGIFAFW AGWTRRWISD DGLIVLRTVR | 60 |
| NLLAGNGPVF NAGERVETNT STLWQYLIYL VALVTDARLE DIALWLAIIF TTAAAVVGVL | 120 |
| GTAHLHRGKI AVLLPAGIIA YFSLSPARDF ATSGLEWGLS LLWIAVQWLL LVLWASTGPA | 180 |
| SGDAKFKANG LFNPGAVTYV LAFWSGLSWL VRPELALYGG LTGVLLLLTA ASRSSVAGIL | 240 |
| AAALPLPLAY QIFRMGYYGL IVPHTAVAKS ASDAMWGTGW DYVEDFTGPY NLWLGLAILL | 300 |
| AAGAVTASML GAAAYGRHLT RTGLRTPGVA VGVLVVCALV HFLYVIRVGG DFMHGRMLLL | 360 |
| PLFAILLPVA VIPVNVVERS WRDAVAGVLV VAVWIYSAVV FVNGHGWENT GEDVVDERDF | 420 |
| WIDFTNRDQD NPPLYAEDFL TVDSMNDFTE VMRDQTLARP TGQQLNILVG TDPNTYSWIT | 480 |
| TPRMEGPEAG DLGNLPPTVF LVNLGMSSMN APLDVRVTDL IGLATPLAAR QPRIEGGRIG | 540 |
| HDKLMGLEWQ VAESATPLAY TPGWIDFDTT YRARQALRHP ELVHLFHVYR EPMSLHRFLA | 600 |
| NIKYALTDGR TLQISNDPED LLKEFDPTPA EFQEGLPQIA WPVDIHLDSP R | 651 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT89 | 26 | 626 | 1.1e-219 | 0.9910071942446043 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMA01273.1 | 0.0 | 20 | 651 | 45 | 678 |
| AGT06584.1 | 0.0 | 20 | 651 | 45 | 678 |
| QDQ23006.1 | 0.0 | 20 | 651 | 45 | 678 |
| ASW15175.1 | 0.0 | 20 | 651 | 45 | 678 |
| QDQ19442.1 | 0.0 | 20 | 651 | 45 | 678 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8NLR0 | 0.0 | 20 | 651 | 45 | 678 | Terminal beta-(1->2)-arabinofuranosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=aftB PE=3 SV=1 |
| O53582 | 2.20e-109 | 40 | 605 | 19 | 594 | Terminal beta-(1->2)-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=aftB PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
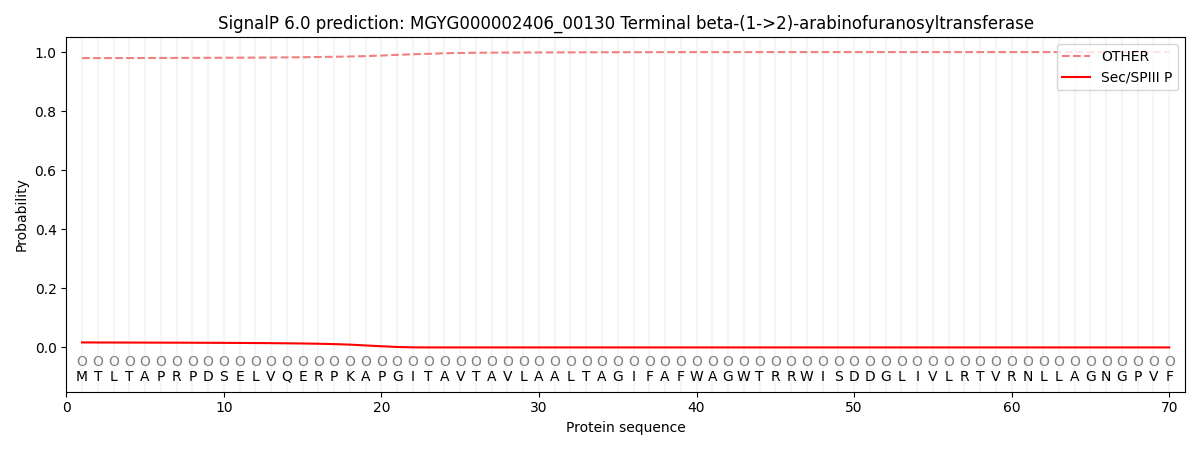
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.980591 | 0.002104 | 0.000073 | 0.000010 | 0.000006 | 0.017251 |
TMHMM Annotations download full data without filtering help
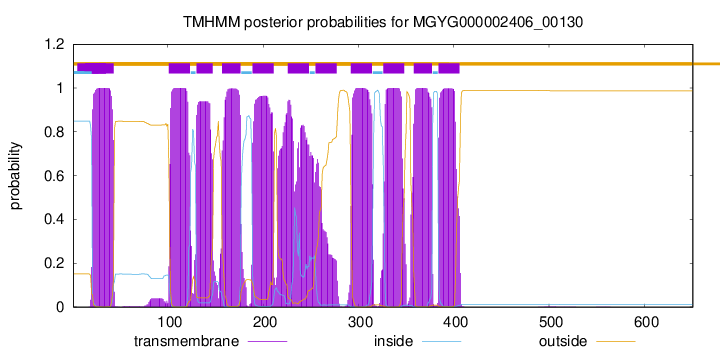
| start | end |
|---|---|
| 21 | 43 |
| 101 | 123 |
| 130 | 147 |
| 157 | 176 |
| 189 | 211 |
| 226 | 248 |
| 255 | 277 |
| 292 | 314 |
| 326 | 348 |
| 358 | 377 |
| 384 | 406 |
