You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001018_00171
You are here: Home > Sequence: MGYG000001018_00171
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900751785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900751785 | |||||||||||
| CAZyme ID | MGYG000001018_00171 | |||||||||||
| CAZy Family | PL8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10460; End: 14674 Strand: - | |||||||||||
Full Sequence Download help
| MKLKKVNRLL ALACAAAVTA SGFCPAAPVK AKETSGIENA EISEQQEESG ISMQAEDSTE | 60 |
| NMILYWSDDL EETTAVSGTA ASYWADGTAA KNWTGLWQAK AFTNEKAVIS LDKETFASGS | 120 |
| QSVHYSSTDA SGRISVSLGA ALQNVDFTKN YILRAKVKAE NVTVSGNNGF YMRGKANNVT | 180 |
| ITPEGTRVNG TTDGWITYDV PLQNLVSVGG AQSGSLALEI FFDYLTGDVW FDSIELWQDY | 240 |
| QISLSETEKT IKPGESFQLE VQCDSEEVDL SKVTWSSSNP DAVSVDENGM ITAEALGTAV | 300 |
| ITAKLDESHT ASCTVQVDDP EMLAPQYAEM RSRWTERLTG NGSSITDDED FQTSMETMAQ | 360 |
| DAEDAMENMA DIPADGSHVD ALWSDLDLEI KYVATSDASY TEDLNTAYTR LQAMATAYAA | 420 |
| ENCRLYHNED LKERILYALE WLYDNGYNEN YNVEKQLYGN WWHWEIGIPQ ALGSTVILMY | 480 |
| EDLTQEQIDK FYATLYRFNQ DPTVVYKVQG WGTMEMTSAN LMDTSLVAAL RSAIGNTQDG | 540 |
| IGAAVNALGT VTGFVTEGDG FYEDGSCIQH SNLAYTGGYG LTLLKGIERI LLLSNDTAWQ | 600 |
| ASADDLESVY TWIWEGYRPL FADGAMMDMV SGRSIARPSH TELETGRGIL EAIVLLADGA | 660 |
| PDDRKEQLLS FATKQVLAGA ENLDTFYSGM EASSMIAAKQ LAADDSVEAD DGTPYTKIFG | 720 |
| SMDKATIHRE GYTLGISMFS SRTGNFEYMN KENTKGWHIS DGALFLYNGD AGQFSNNYWN | 780 |
| TIDPHRLPGI TTDHTEGTNY ESGLAYTSDK DYAGGSSVED LYATIAMDFH GQNTDLTAKK | 840 |
| AWFAFDDEVV ALGTDISGIT KDTETIIENK QIRDDGSNAL VVDGEETQAE LGESSAAGVE | 900 |
| YAWIEGNSGT DSIGYYFPEG EDLEIKREAR TGSFQDINGA VADGAAGSED VTRDYLSLAV | 960 |
| SHGDGTEEGA EDYAYVLLPG RTEEETAEYA SGSEIEIISN TAEVQAAADR SSGASGYQFW | 1020 |
| TAASAGNVSA DQASSVTMKE EDGTLKLGIS DPSQTQDSVT IHVSGYKNLQ YVDGDQEAVV | 1080 |
| NATEDGVDIT VDTSAGAGKT YELTLTYEEE GAETSVSTAL LGYAIELAKD ADLTDVVPAV | 1140 |
| VEKFNAALSN AEEIVEKVKA GDTSITQEMM DESWQELVRA MQYLSFKQGD KTNLNKVIAM | 1200 |
| AESLDLDGYL LAGQDVFTEA LDSARAVAAD ENAMEKEIEE AWRELLAAMG NLRLKPSKEL | 1260 |
| LEDLISQAQS VDTAVYTEES VQALTAALAD AVAVFDDDQA DKEQVEEAQS ALKAALAGLT | 1320 |
| AKTEAGGSDA AGADTDTGNS GTGSAGSGTA GTVNSSADRA VKTGDSSPAV PLYAAAAALA | 1380 |
| LVCAAGSGLG KQADRGRKES KDKE | 1404 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL8 | 717 | 983 | 1.3e-78 | 0.9884169884169884 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01083 | GAG_Lyase | 0.0 | 327 | 1052 | 2 | 693 | Glycosaminoglycan (GAG) polysaccharide lyase family. This family consists of a group of secreted bacterial lyase enzymes capable of acting on glycosaminoglycans, such as hyaluronan and chondroitin, in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. These are broad-specificity glycosaminoglycan lyases which recognize uronyl residues in polysaccharides and cleave their glycosidic bonds via a beta-elimination reaction to form a double bond between C-4 and C-5 of the non-reducing terminal uronyl residues of released products. Substrates include chondroitin, chondroitin 4-sulfate, chondroitin 6-sulfate, and hyaluronic acid. Family members include chondroitin AC lyase, chondroitin abc lyase, xanthan lyase, and hyalurate lyase. |
| pfam08124 | Lyase_8_N | 6.37e-83 | 334 | 671 | 1 | 320 | Polysaccharide lyase family 8, N terminal alpha-helical domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| pfam02278 | Lyase_8 | 1.82e-75 | 716 | 980 | 1 | 250 | Polysaccharide lyase family 8, super-sandwich domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| pfam02884 | Lyase_8_C | 8.28e-13 | 998 | 1060 | 1 | 67 | Polysaccharide lyase family 8, C-terminal beta-sandwich domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| pfam02368 | Big_2 | 8.57e-10 | 241 | 315 | 2 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SQI53822.1 | 1.87e-173 | 106 | 1103 | 70 | 1046 |
| SYX85126.1 | 9.65e-170 | 64 | 1130 | 33 | 1095 |
| AZS13398.1 | 5.85e-165 | 68 | 1112 | 48 | 1068 |
| AYV66165.1 | 5.95e-162 | 79 | 1103 | 49 | 1048 |
| QJX62049.1 | 7.38e-162 | 79 | 1103 | 44 | 1043 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1J0M_A | 1.21e-120 | 326 | 1101 | 3 | 745 | CrystalStructure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan [Bacillus sp. GL1],1J0N_A Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan [Bacillus sp. GL1] |
| 2E24_A | 1.67e-120 | 326 | 1101 | 3 | 745 | ChainA, Xanthan lyase [Bacillus sp. GL1] |
| 2E22_A | 8.49e-120 | 326 | 1101 | 3 | 745 | Crystalstructure of xanthan lyase in complex with mannose [Bacillus sp. GL1] |
| 1X1H_A | 5.96e-119 | 326 | 1101 | 3 | 745 | ChainA, xanthan lyase [Bacillus sp. GL1],1X1I_A Chain A, xanthan lyase [Bacillus sp. GL1],1X1J_A Chain A, xanthan lyase [Bacillus sp. GL1] |
| 6F2P_A | 3.41e-115 | 324 | 1103 | 3 | 747 | Structureof Paenibacillus xanthan lyase to 2.6 A resolution [Paenibacillus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9AQS0 | 2.74e-117 | 326 | 1110 | 28 | 779 | Xanthan lyase OS=Bacillus sp. (strain GL1) OX=84635 GN=xly PE=1 SV=1 |
| Q54873 | 2.91e-92 | 145 | 1013 | 121 | 950 | Hyaluronate lyase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0314 PE=1 SV=2 |
| Q53591 | 9.99e-74 | 59 | 1012 | 9 | 930 | Hyaluronate lyase OS=Streptococcus agalactiae serotype III (strain NEM316) OX=211110 GN=hylB PE=1 SV=2 |
| Q59801 | 1.04e-73 | 315 | 980 | 37 | 692 | Hyaluronate lyase OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=hysA PE=3 SV=1 |
| P0CZ01 | 1.05e-53 | 291 | 1105 | 11 | 796 | Hyaluronate lyase OS=Cutibacterium acnes (strain DSM 16379 / KPA171202) OX=267747 GN=PPA0380 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
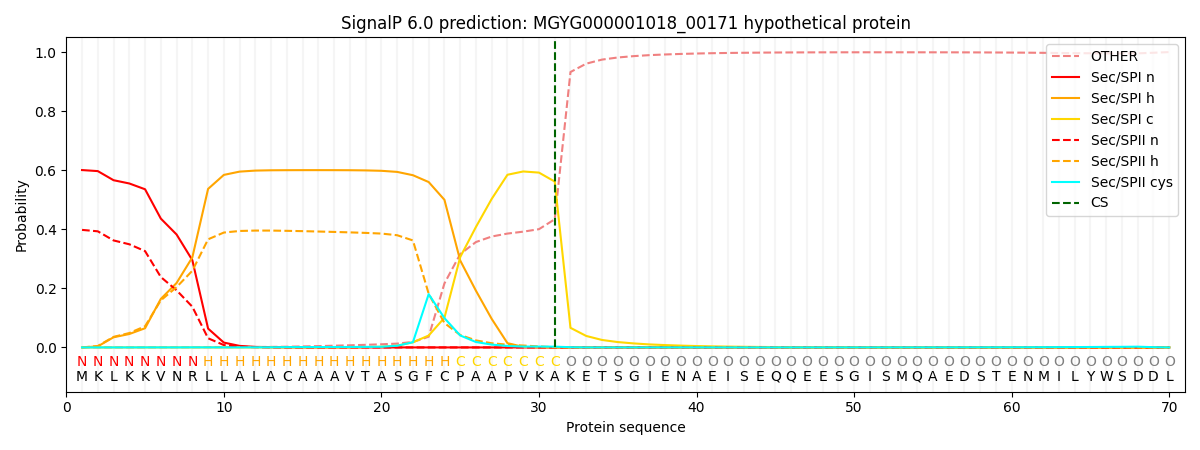
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001345 | 0.587641 | 0.408332 | 0.001905 | 0.000447 | 0.000284 |
