You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000014_00174
You are here: Home > Sequence: MGYG000000014_00174
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
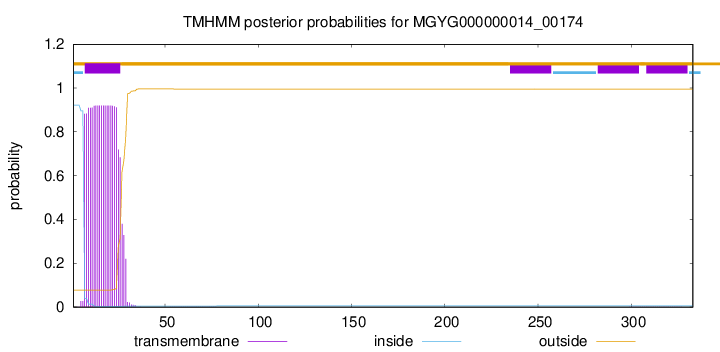
TMHMM annotations
Basic Information help
| Species | Clostridium butyricum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium butyricum | |||||||||||
| CAZyme ID | MGYG000000014_00174 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 166708; End: 167709 Strand: + | |||||||||||
Full Sequence Download help
| MKKVRNLCLI AALSLIAVGG SSFYQIPVNA ATTIEVSNSG TSLKDALAKA KSGDIVNITG | 60 |
| TVKSGTVNVP AGVTITGEKG KSKIDFSSTS GSSGKGFVIK SDGASIKNLD VYGAADNGIY | 120 |
| IEGSKNNITN VNVYKNKDAG VQLSNGAANN TLTKVYSYSN ADQTGENADG FAIKLHSGEG | 180 |
| NKLIECTAEG NSDDGYDLYA AHGAVTFIRC KAINNGNCDG IKGDGNGFKL GGVDNKTSGV | 240 |
| AAHLDPLNHE LTDCIAIGNT GSGFDRNNQN GVVKMTNCTG ENNGEYNFNF PLKGKPSALG | 300 |
| YEVTFGKAIM NGCTSINGGN VITGASLTDC TGF | 333 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 37 | 305 | 1.3e-67 | 0.7032085561497327 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 3.97e-05 | 97 | 279 | 4 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSX04214.1 | 3.02e-200 | 1 | 333 | 1 | 333 |
| QJU44567.1 | 3.02e-200 | 1 | 333 | 1 | 333 |
| ALS17289.1 | 3.02e-200 | 1 | 333 | 1 | 333 |
| AXB85212.1 | 3.02e-200 | 1 | 333 | 1 | 333 |
| QCJ07648.1 | 3.02e-200 | 1 | 333 | 1 | 333 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 8.44e-18 | 157 | 290 | 170 | 298 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A7 | 5.30e-17 | 157 | 290 | 195 | 323 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 5.87e-16 | 157 | 293 | 195 | 328 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P22751 | 8.10e-13 | 157 | 302 | 526 | 655 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
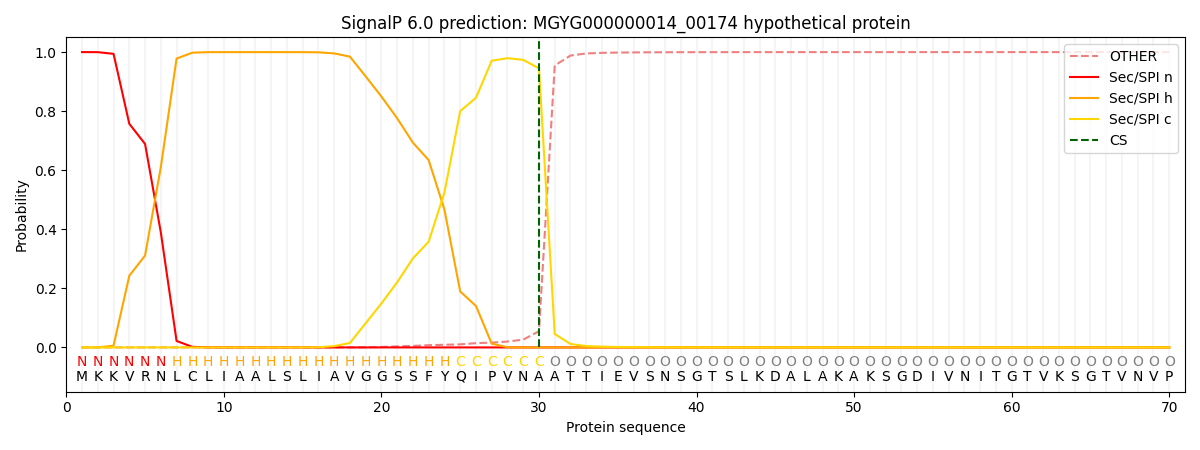
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000313 | 0.998950 | 0.000210 | 0.000169 | 0.000164 | 0.000152 |