You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000967_00062
You are here: Home > Sequence: MGYG000000967_00062
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
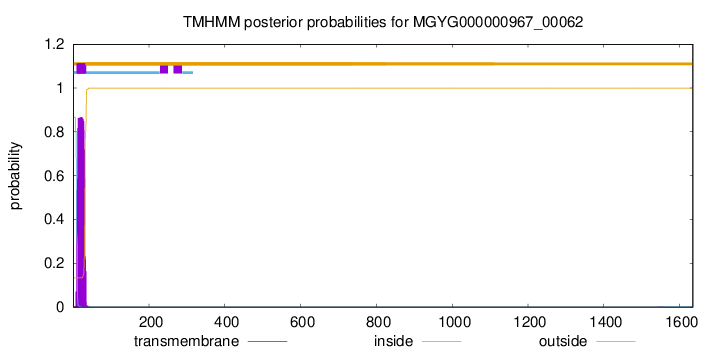
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900543365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900543365 | |||||||||||
| CAZyme ID | MGYG000000967_00062 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19386; End: 24293 Strand: + | |||||||||||
Full Sequence Download help
| MKKCLKKTLS GILAATMLFS SVTVSNVFAA PSFSQVGGWF ESIYAEIPSI KDADVTGVSY | 60 |
| SGATSGTLSG DDLTYLVRDM SGGVRIDVPG LKAGTYTLTV TTSSGTVTKD NIIVQEYDRS | 120 |
| GYAHYNYTEG VGAYNDDGTV KANAKILYVT NENKDTVSVT SKDGTTVTGI GNILNSAGKD | 180 |
| AGTGTTSKGG KPNTNAGIIG KLADDGTPLV VRIVGDVKAP AGVTAYDSVD YGGTVGDNGY | 240 |
| MARMQGGKDV TIEGIGADSS INGWGIHFIC DTVGYSKGNG KSFEVRNIKF QNVPEDCVGM | 300 |
| EGQQEGSTIT AGVERCWVHN CSFYAPSISN PAESDKSGGD GACDFKRGQY FTNSYCYYKG | 360 |
| YHKTNLVGSS DSSLQFNLTY HHNYWQNCEA RGPLGRQANI HMYNNYYEGQ TDYAMNTRAN | 420 |
| CYIFSEYNLF YMCKNPQRVD AGAIKSYNDS LSGCIEAMEA KVVTDKSTKV SSGCKYENFD | 480 |
| TDSSLSYIPS NNYKLDTSIV SAKKNIMAYA GTMKENPVTA ESVNTSIVLS DRQPTASVQL | 540 |
| PYDKDLNSSY VTNKSGAIDN IVFNVGKTAV DSISTATDTN GQNIVFYVNE PVNISITDGG | 600 |
| ATYPVILLND SGEAIITGTG TANNVPAGTY FIQSSGFQPG KSGSPAKFKE AKITHLSIVS | 660 |
| AGSELPTIAE PTTSEPTTSD SSNPTEGTTS NQEPDTEETT SKPYDGEGLV WNYTSGENTL | 720 |
| GVNFNGNDYT GSSVTYNGST FTKAAKMESS TSIGFTAPGS GKLTIVSKSS KNPATIKVNG | 780 |
| ETVTISQDGA TTIDVPAGAV NITKGTTSTY LYLLEFKGDS VETTTNVTTV TEATTEATTK | 840 |
| AEATTEATTE ATTVPEGVVV SVGSTTAKTG DKITVPVKLT GLNTLENYAV TVNYDSNVLT | 900 |
| LSDVVSFVDS NNFVVNKNNA GVIKVAYAND NAGSDDSFNG DVLFSMTFTV KSENDTTSSI | 960 |
| NATVNQLNDG VTATVVNGTV TVDNAVTPVS IPGDVNKDGV VNSVDAAIVL KIASGIITDT | 1020 |
| TPYDMVAADC DGKFGIDGRD VIWILNHQTT DDNTSTTEAT TEETTSKTED TTKAEATTES | 1080 |
| TTTSPVVTDG LPAGSYDLTA SVTDKIDTTN ARDVSTAGIK LRSENYIEFV AGVSGKLTLT | 1140 |
| VSGKAAKLVS VANGAETEVA LNSNSANVTA GTTYRVYGTE SGSNTTITNL VLASDGTVVE | 1200 |
| PTTQATTNSE VTTSKTETTT KTSGGDATTE TTTTDLSGAI NISAGDSASL ATALKNATAG | 1260 |
| TTINLAPGTY KMSAATSMSK SGTASKPITV TCANGMATLD YDGTSGRAIT AKADYINFSN | 1320 |
| LTVKNGGDNG MYITGGHINV ENCIFQANGD TGLQISGGGN NVLVKNCTSF DNLQTENADG | 1380 |
| FAAKLGAGEN VVFDGCIAYC NSDDGWDLFS KSGDQQNKYP ITLRNCIAFK NGQLTDGTVE | 1440 |
| KSGDRNGFKL GGGGYGAAHI VENCIAFDNG ACGFTDNNNP SLAVLKNCTG YSNATADVKK | 1500 |
| HNFSIYRATE GIEVTNCLSY VLNADPNGDG KDRFDGSSSG TTYNANATVA NSVFGCANKY | 1560 |
| YKIAGATKIT ATTQLATNGT EVTVSDSDFQ TLELPYTDIL KVHEQMRNAD GSIKLNGFLQ | 1620 |
| PKAGTDIEGM GAQFN | 1635 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 1247 | 1628 | 5.5e-93 | 0.9064171122994652 |
| PL1 | 242 | 431 | 9.2e-44 | 0.8316831683168316 |
| CBM77 | 722 | 808 | 1.3e-17 | 0.8252427184466019 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.49e-26 | 246 | 436 | 99 | 279 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.73e-26 | 247 | 430 | 15 | 186 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 5.04e-16 | 208 | 430 | 1 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| pfam18283 | CBM77 | 6.76e-16 | 733 | 813 | 29 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| cd08548 | Type_I_cohesin_like | 9.36e-12 | 858 | 995 | 1 | 135 | Type I cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I cohesins; their interactions with dockerin mediate assembly of a range of dockerin-borne enzymes to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU23076.1 | 4.30e-219 | 35 | 525 | 760 | 1249 |
| ACR71161.1 | 7.19e-206 | 32 | 820 | 41 | 896 |
| BBF42492.1 | 2.42e-141 | 3 | 671 | 2 | 668 |
| CDR31241.1 | 9.24e-117 | 35 | 635 | 332 | 894 |
| ADZ82421.1 | 1.49e-68 | 1 | 529 | 1 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VMV_A | 1.82e-19 | 248 | 430 | 79 | 246 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1RU4_A | 1.59e-16 | 1260 | 1493 | 48 | 292 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 2QX3_A | 6.44e-13 | 243 | 436 | 67 | 252 | Structureof pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris],2QX3_B Structure of pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris] |
| 5AMV_A | 1.51e-12 | 249 | 516 | 128 | 399 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
| 1BN8_A | 1.71e-12 | 249 | 516 | 149 | 420 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 6.10e-18 | 1249 | 1493 | 62 | 317 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.08e-15 | 1260 | 1493 | 73 | 317 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| Q65DC2 | 1.99e-15 | 248 | 517 | 108 | 341 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
| B1B6T1 | 1.99e-15 | 248 | 517 | 108 | 341 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
| Q8GCB2 | 1.99e-15 | 248 | 517 | 108 | 341 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
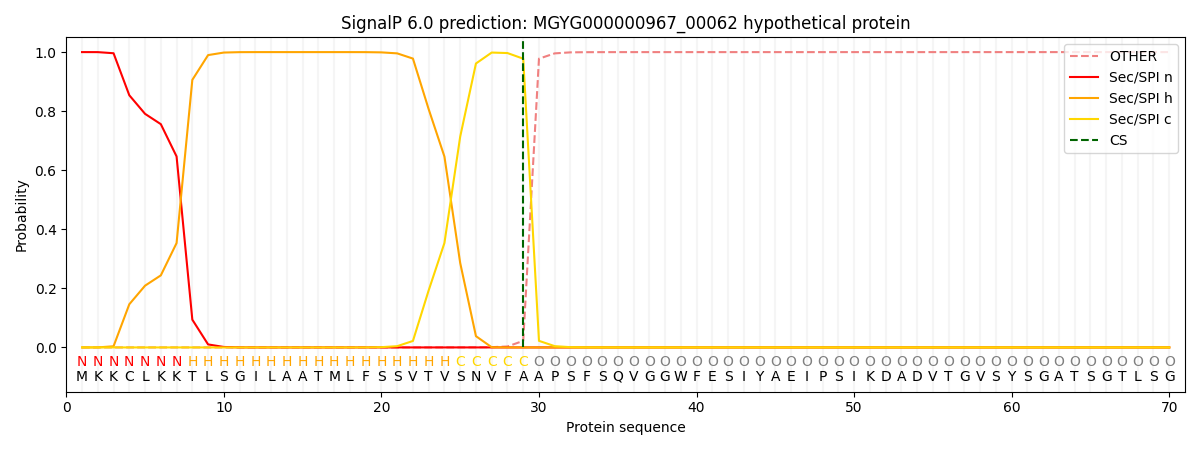
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000216 | 0.999130 | 0.000165 | 0.000174 | 0.000153 | 0.000140 |