You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001160_00198
You are here: Home > Sequence: MGYG000001160_00198
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
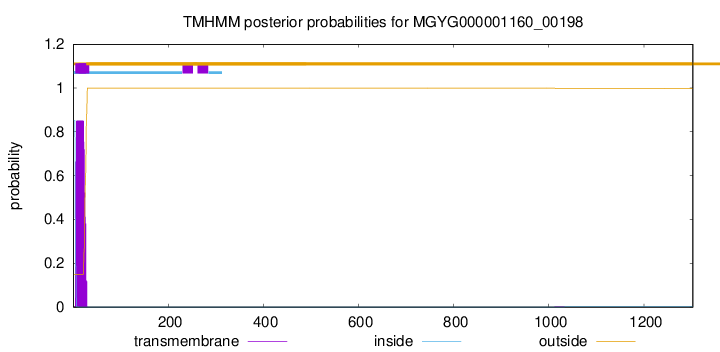
TMHMM annotations
Basic Information help
| Species | CAG-274 sp000432155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; CAG-274; CAG-274 sp000432155 | |||||||||||
| CAZyme ID | MGYG000001160_00198 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 207883; End: 211794 Strand: + | |||||||||||
Full Sequence Download help
| MKKRLVCFGL ALAFSLSSFS AISFAEGSWK KGWTGKADAA AYTVNANGST VTMSNDNVNN | 60 |
| GKFTDGEDSI VYYANELKAD EDFELKATVN IDSYNTAKDG SNPQQGSVGI GVLDSLYNKT | 120 |
| DDKTYDDGVF LGSYASKKDS PMAIHSLVRA NSSKKTVGDA LSDTFANTGE NLGTFDLSIK | 180 |
| KVGNAYTLTC GDKSTTVEMT AMEDEIYPCL YIARNAKATF TNVSLKTETR KATSLTLKGK | 240 |
| CKTAYSYGEK LDLSGLTGTV TYDDGTKEDV TGFLASGYDA TKTGKQTITL SYGEAKANID | 300 |
| VSVANVALTD VKIGFTPVKT DYSLNGPFSS TGISVEAVYS DGTTKKLDSD QYTFSLNGKT | 360 |
| IKDGDAVTGI GHQIVKVNVK PQKGINTTAV GSYNIYINKN AYSSLEAKGK GFDTEYYLND | 420 |
| QFDTNTIEVT ASYTNDSGKV VSEIVPAGSY TLTGFDSKTV GEKEVKVSAG GLDVSNTVTV | 480 |
| KEKQVTGIEL TKYPKTTYNV GESFDKEDMA VVVKYDNGDE DELTDYTVNT DKFDTSKVGK | 540 |
| TSVTVSSKEG SVELPCTVVE AKDAKWRKVV FGQSSNYDKQ DEGISGVTAD DYGTVNGKIN | 600 |
| VRSWNGSGKI TQDHDGISYY YTTVDGNEDF RVTADIKVNK YLEHDNNDTK RSGQEAFGIM | 660 |
| ARDVIPLTGE DGKIVTDESK AKKDSEGVSQ PLDNGAVFAS NMVICGGYSG TGYPADKSTK | 720 |
| DYDKIINMNR INLIVREGVT ATDGGGKRIG PYAINSEFPK EGNTYRVTLE RMNGGLYAKC | 780 |
| YDYQTGKTQT TVYNDDSFLT VQSDKVYVGF FTSRWADIDV SNVEFRTSDR STDQKVAEKE | 840 |
| EEPATAGISF KTKGYSTSEN YGFDLATSDS KGKVTIKVND AVVAQNVDMA DTQHFTAKLK | 900 |
| NDTANKIVAV YTPDKSLNLS SYEPIVVRKT VYCKETPANA TTLYVSPQGS FNGDGTKENP | 960 |
| FDIDTAIGLC KAGQTIQLAG GVYNRTSRID ISIGDDGTAE KPKTIRADEN DRAIIDLGAT | 1020 |
| CAGFYTGGNY WVFENMDVRN SGNNLKAFNL GGSHCVVRNC RFYDNRDTGF QISRIDDDQT | 1080 |
| REFWPGYNLI ENCESFNNCD PAMINADGFG TKLTSGDGNV YRNCKSHHNL DDGWDCYTKV | 1140 |
| NTGAIGAVTL ENCQAYKMGL RLNEDGTETP YGAGGHNGFK MGGENIAVNH VLKNCVAYDN | 1200 |
| LACGVTTNFN PSLTLIDVKA YNNAESNFNF FTDKADQYNY TVTGAVSYNG GAVDRIPTEN | 1260 |
| YNKEYKNNSQ TPLISDSNYW ELALGKSVNK SGKEANESML KMK | 1303 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 939 | 1283 | 2.2e-103 | 0.9146666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07523 | Big_3 | 1.82e-07 | 242 | 303 | 4 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| cd14251 | PL-6 | 2.33e-07 | 965 | 1093 | 9 | 133 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam07523 | Big_3 | 7.44e-07 | 492 | 558 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam14592 | Chondroitinas_B | 2.30e-04 | 959 | 1093 | 3 | 134 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
| pfam07523 | Big_3 | 0.003 | 413 | 480 | 4 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANW97669.1 | 1.95e-220 | 44 | 1297 | 59 | 1222 |
| BAC87940.1 | 7.60e-220 | 44 | 1297 | 59 | 1222 |
| AGI38277.1 | 1.50e-219 | 44 | 1297 | 59 | 1222 |
| AGC67199.1 | 1.50e-219 | 44 | 1297 | 59 | 1222 |
| ANX00231.1 | 2.11e-219 | 44 | 1297 | 59 | 1222 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.79e-27 | 944 | 1223 | 18 | 292 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 1.33e-11 | 942 | 1229 | 4 | 324 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 2.01e-57 | 566 | 1261 | 34 | 686 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 3.05e-28 | 944 | 1282 | 43 | 371 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.43e-26 | 944 | 1223 | 43 | 317 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
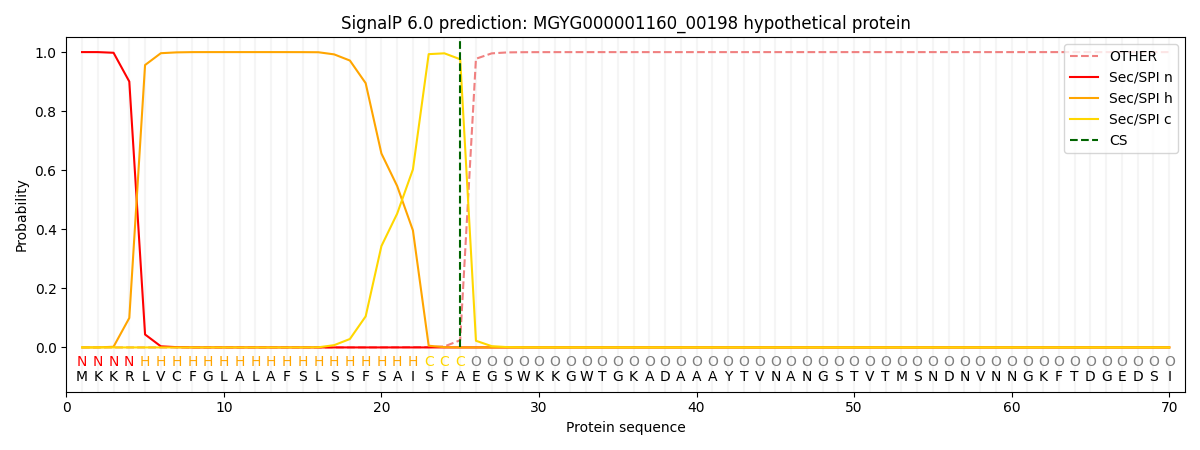
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000247 | 0.999050 | 0.000209 | 0.000179 | 0.000163 | 0.000146 |