You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002638_00200
You are here: Home > Sequence: MGYG000002638_00200
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoanaerobaculum orale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoanaerobaculum; Lachnoanaerobaculum orale | |||||||||||
| CAZyme ID | MGYG000002638_00200 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28865; End: 33451 Strand: + | |||||||||||
Full Sequence Download help
| MKRNILRCFM AYILSILLLS NNLEVFASTL TNGIEITSET EERSDTVISK ATDSNAKEWE | 60 |
| FTYFGPSTST ITNTVNDDAD IDGEIILKSA TYKKDGSIDK KGGKFVSSDP ADGISFYYTK | 120 |
| IDPSKENFIL EADVRIDYMN PKPDGQEGFA LMVRDSIGEK GDPASSESNL VSIGATQLPK | 180 |
| GELNSTSQVK DMLGVRAYTG IDNKIKPNPD TFKVYRLGFD KDGGKVSSGD TYRVRLEKNS | 240 |
| SSYVVYQLAM DGSELYKYEI PIQAKDIRAN SISGYEELND PMNVQDRFAY VGFAVARGMN | 300 |
| VTYSNIKFEK SEFDANDWRP QESRYIDLQA KITSPDTAAE ENYTLVFKAN ANGSAKIYQD | 360 |
| NVLKDSVEVV NNEELKKDYD LSAGKNFSVE FTPEAGYEPE PFVKLKSYET VTLNKSVSWR | 420 |
| TIGKDSYIYV SNTGLDVNDG SGYDNAISLE SAIKYAKPGQ TILLKPEVYD MSNKSLIVQK | 480 |
| GRNGREDNPI RIQGDGGFAT LDFKRSGKGF SLGGDYWELK QFNITNTADG YKGLGISGSH | 540 |
| NIAERLNLFN NGTTGLQISG SSEDSKDKWP SYNYILNCTS MNNADSAMED ADGFAAKITS | 600 |
| GVGNVFDGCI AAYNADDGWD LFAKVATGSI GDVTIKNSVA YRNGYILIKN GGNKKNFEFA | 660 |
| PGTVDEYGNV SFDKPNIMLD AGNGNGFKMG GSNVSGAHKL DNSISYENKA KGIDSNSGPN | 720 |
| IKISNSTSYN NGSYNVALYT SDKTLTTDFS AKGILSFRKE TNIGEQLSLQ NQQSSDVMDE | 780 |
| TTFYWNKDEN ISKNTASKVI TMDENDFINL DTENDPKRNE DGKINMNGLL LLKPEAKEKL | 840 |
| SSGAGGRAFG EKENIATVWA VGDSTVSGFN DNYYIPRVGY GEELKNYLNA EVYNLAVSGA | 900 |
| SSKDFTTMKN YDVLLNGNSD VPKLGDGDNE KFLIIGFGHN DEKSELSRYT DPNSSYENEG | 960 |
| SFANSVYKNY VKPALERGVT PIIVTPIARL TDDNTSESYN SPSGHITNDV VVGNNIFKGG | 1020 |
| DYRKALIDMS NQLGLEYIDL TEKTIDLNIE MGNDARYLYA YTGAKKEGDN LVPKGLDKTH | 1080 |
| TNLYGAKRNA YLIANADTTL KKYSKKREYP VKEDFFEDAV NKDYKVIDYI TPTEESKNWP | 1140 |
| EFLDVVGNVF TGSVFGNVGG EDKITGGDFE VFKSDKSINL TVKNNRGKIA SNVDGIMMYY | 1200 |
| KRLPENTEFT LKAKVKINDI AKNDQVSFGL MARDDMYVDR SITDSIGDYV VAGSRMQGKI | 1260 |
| NGFGRKNSKL YDGGVANIIY QKGDELDLTL TKTQDGYMVK YGENEPVSAG FDYPLTQIDS | 1320 |
| EYIYVGFFVA RNANITFSDV SLKLKDNVDN EKNKAGEKIV PDTPSKNNNI TEEKPKDGLP | 1380 |
| NDSKKDQNTV ENHHQEVAEH SKVSDNKNKP ASDDSRVVIP KNSNSKAKNR RSSSSSRSSS | 1440 |
| KGSKKVFHNT FTGWKPETVD GHNVWKYVEN GKNVTSQWAY IFNPYTNRNA WFKFDKDGIM | 1500 |
| ETGWINENGI NYYLSEISDG SLGEWIKK | 1528 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 426 | 836 | 9.1e-107 | 0.9786666666666667 |
| CE12 | 857 | 1095 | 7.3e-39 | 0.9857142857142858 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01821 | Rhamnogalacturan_acetylesterase_like | 1.08e-33 | 857 | 1094 | 2 | 195 | Rhamnogalacturan_acetylesterase_like subgroup of SGNH-hydrolases. Rhamnogalacturan acetylesterase removes acetyl esters from rhamnogalacturonan substrates, and renders them susceptible to degradation by rhamnogalacturonases. Rhamnogalacturonans are highly branched regions in pectic polysaccharides, consisting of repeating -(1,2)-L-Rha-(1,4)-D-GalUA disaccharide units, with many rhamnose residues substituted by neutral oligosaccharides such as arabinans, galactans and arabinogalactans. Extracellular enzymes participating in the degradation of plant cell wall polymers, such as Rhamnogalacturonan acetylesterase, would typically be found in saprophytic and plant pathogenic fungi and bacteria. |
| NF033840 | PspC_relate_1 | 0.002 | 1452 | 1527 | 550 | 615 | PspC-related protein choline-binding protein 1. Members of this family share C-terminal homology to the choline-binding form of the pneumococcal surface antigen PspC, but not to its allelic LPXTG-anchored forms because they lack the choline-binding repeat region. Members of this family should not be confused with PspC itself, whose identity and function reflect regions N-terminal to the choline-binding region. See Iannelli, et al. (PMID: 11891047) for information about the different allelic forms of PspC. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL00444.1 | 0.0 | 50 | 1342 | 32 | 1327 |
| QPL60595.1 | 4.00e-129 | 50 | 838 | 83 | 859 |
| AYQ60810.1 | 7.60e-129 | 50 | 838 | 83 | 859 |
| AWX46906.1 | 7.60e-129 | 50 | 838 | 83 | 859 |
| AYA33762.1 | 7.60e-129 | 50 | 838 | 83 | 859 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.17e-34 | 429 | 732 | 18 | 293 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 5.87e-10 | 571 | 716 | 168 | 303 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 2.10e-42 | 74 | 735 | 69 | 643 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 3.47e-36 | 429 | 732 | 43 | 318 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.03e-33 | 429 | 732 | 43 | 318 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| O31528 | 1.35e-13 | 858 | 1094 | 5 | 199 | Probable rhamnogalacturonan acetylesterase YesY OS=Bacillus subtilis (strain 168) OX=224308 GN=yesY PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
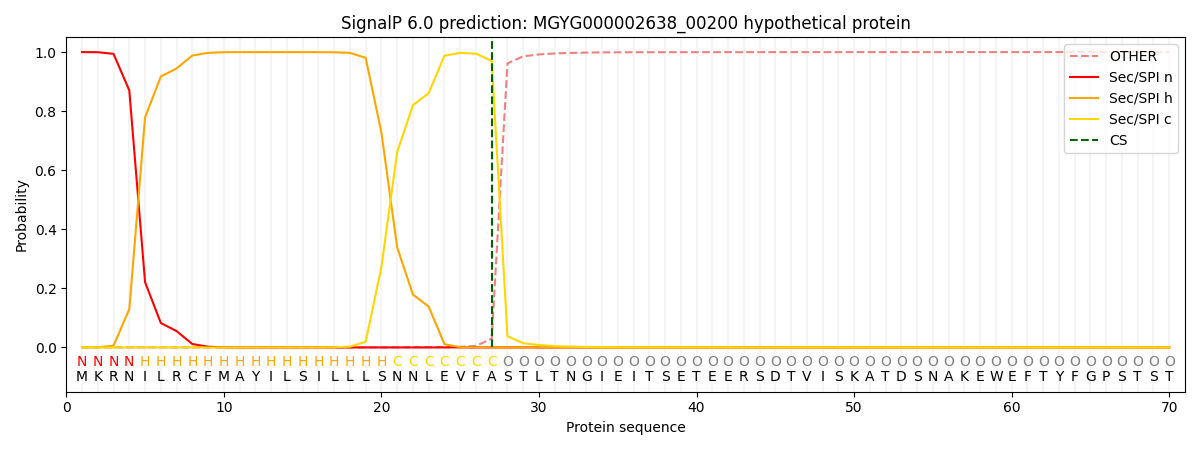
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000392 | 0.998855 | 0.000232 | 0.000178 | 0.000164 | 0.000152 |
