You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000003_00272
You are here: Home > Sequence: MGYG000000003_00272
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes shahii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes shahii | |||||||||||
| CAZyme ID | MGYG000000003_00272 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 299735; End: 302452 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 671 | 2.3e-75 | 0.6768617021276596 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.75e-22 | 29 | 466 | 14 | 446 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.32e-21 | 18 | 449 | 2 | 443 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 2.89e-11 | 198 | 305 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 2.12e-09 | 95 | 474 | 113 | 506 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 9.13e-09 | 95 | 428 | 124 | 462 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK63994.1 | 0.0 | 1 | 905 | 1 | 905 |
| QPH40113.1 | 0.0 | 23 | 904 | 24 | 906 |
| AZI26708.1 | 0.0 | 23 | 904 | 24 | 906 |
| QIH37197.1 | 0.0 | 28 | 904 | 20 | 896 |
| QMV71213.1 | 0.0 | 28 | 904 | 20 | 896 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GM8_A | 5.83e-21 | 34 | 466 | 13 | 436 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 5EUV_A | 4.26e-16 | 58 | 454 | 27 | 391 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d],5EUV_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d],5LDR_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 5LDR_A | 4.26e-16 | 58 | 454 | 28 | 392 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 6U7I_A | 7.76e-16 | 81 | 428 | 54 | 412 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 6HPD_A | 6.18e-15 | 33 | 504 | 43 | 509 | Thestructure of a beta-glucuronidase from glycoside hydrolase family 2 [Formosa agariphila KMM 3901] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6LL68 | 7.55e-16 | 119 | 459 | 149 | 488 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| Q7MG04 | 8.82e-15 | 119 | 428 | 150 | 457 | Beta-galactosidase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=lacZ PE=3 SV=1 |
| Q8D4H3 | 8.83e-15 | 119 | 428 | 150 | 458 | Beta-galactosidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=lacZ PE=3 SV=2 |
| T2KN75 | 3.37e-14 | 33 | 504 | 34 | 500 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| T2KPJ7 | 6.91e-14 | 93 | 435 | 105 | 437 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
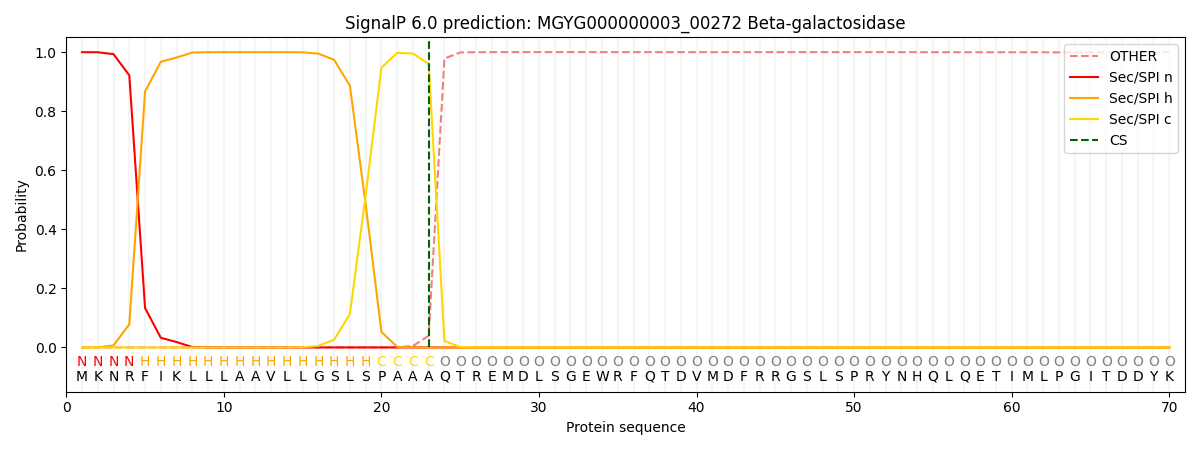
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001880 | 0.993425 | 0.003205 | 0.000483 | 0.000464 | 0.000498 |
