You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000003_01223
You are here: Home > Sequence: MGYG000000003_01223
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes shahii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes shahii | |||||||||||
| CAZyme ID | MGYG000000003_01223 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 567949; End: 569463 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 75 | 498 | 1.3e-148 | 0.9904076738609112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 1.19e-56 | 78 | 498 | 36 | 428 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02055 | Glyco_hydro_30 | 2.59e-33 | 89 | 393 | 1 | 313 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam17189 | Glyco_hydro_30C | 1.09e-19 | 434 | 496 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
| pfam02057 | Glyco_hydro_59 | 1.41e-07 | 90 | 354 | 2 | 228 | Glycosyl hydrolase family 59. |
| pfam14587 | Glyco_hydr_30_2 | 9.20e-06 | 75 | 284 | 1 | 224 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK62918.1 | 0.0 | 1 | 504 | 1 | 504 |
| QRO16282.1 | 1.94e-181 | 44 | 498 | 21 | 478 |
| ABR43532.1 | 1.94e-181 | 44 | 498 | 21 | 478 |
| QUR49687.1 | 3.89e-181 | 48 | 498 | 25 | 478 |
| QUT22389.1 | 3.89e-181 | 48 | 498 | 25 | 478 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5NGK_A | 5.52e-178 | 45 | 498 | 34 | 490 | Theendo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
| 2WNW_A | 1.14e-36 | 78 | 498 | 34 | 442 | Thecrystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],2WNW_B The crystal structure of SrfJ from salmonella typhimurium [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 3KE0_A | 5.80e-33 | 43 | 498 | 32 | 494 | ChainA, Glucosylceramidase [Homo sapiens],3KE0_B Chain B, Glucosylceramidase [Homo sapiens],3KEH_A Chain A, Glucocerebrosidase [Homo sapiens],3KEH_B Chain B, Glucocerebrosidase [Homo sapiens] |
| 2WKL_A | 7.90e-33 | 43 | 498 | 32 | 494 | Velaglucerasealfa [Homo sapiens],2WKL_B Velaglucerase alfa [Homo sapiens],5LVX_A Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator [Homo sapiens],5LVX_B Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator [Homo sapiens],5LVX_C Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator [Homo sapiens],5LVX_D Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator [Homo sapiens],6TJK_AAA Chain AAA, Lysosomal acid glucosylceramidase [Homo sapiens],6TJK_BBB Chain BBB, Lysosomal acid glucosylceramidase [Homo sapiens],6TJQ_BBB Chain BBB, Glucosylceramidase [Homo sapiens],6TN1_AAA Chain AAA, Lysosomal acid glucosylceramidase [Homo sapiens],6YTR_AAA Chain AAA, Lysosomal acid glucosylceramidase [Homo sapiens],6YTR_BBB Chain BBB, Lysosomal acid glucosylceramidase [Homo sapiens],6Z3I_BBB Chain BBB, Lysosomal acid glucosylceramidase [Homo sapiens],7NWV_AAA Chain AAA, Lysosomal acid glucosylceramidase [Homo sapiens],7NWV_BBB Chain BBB, Lysosomal acid glucosylceramidase [Homo sapiens] |
| 1OGS_A | 7.90e-33 | 43 | 498 | 32 | 494 | humanacid-beta-glucosidase [Homo sapiens],1OGS_B human acid-beta-glucosidase [Homo sapiens],1Y7V_A Chain A, Glucosylceramidase [Homo sapiens],1Y7V_B Chain B, Glucosylceramidase [Homo sapiens],2F61_A Crystal structure of partially deglycosylated acid beta-glucosidase [Homo sapiens],2F61_B Crystal structure of partially deglycosylated acid beta-glucosidase [Homo sapiens],2J25_A Partially deglycosylated glucoceramidase [Homo sapiens],2J25_B Partially deglycosylated glucoceramidase [Homo sapiens],2NSX_A Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease [Homo sapiens],2NSX_B Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease [Homo sapiens],2NSX_C Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease [Homo sapiens],2NSX_D Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease [Homo sapiens],2NT0_A Acid-beta-glucosidase low pH, glycerol bound [Homo sapiens],2NT0_B Acid-beta-glucosidase low pH, glycerol bound [Homo sapiens],2NT0_C Acid-beta-glucosidase low pH, glycerol bound [Homo sapiens],2NT0_D Acid-beta-glucosidase low pH, glycerol bound [Homo sapiens],2NT1_A Structure of acid-beta-glucosidase at neutral pH [Homo sapiens],2NT1_B Structure of acid-beta-glucosidase at neutral pH [Homo sapiens],2NT1_C Structure of acid-beta-glucosidase at neutral pH [Homo sapiens],2NT1_D Structure of acid-beta-glucosidase at neutral pH [Homo sapiens],3GXD_A Crystal structure of Apo acid-beta-glucosidase pH 4.5 [Homo sapiens],3GXD_B Crystal structure of Apo acid-beta-glucosidase pH 4.5 [Homo sapiens],3GXD_C Crystal structure of Apo acid-beta-glucosidase pH 4.5 [Homo sapiens],3GXD_D Crystal structure of Apo acid-beta-glucosidase pH 4.5 [Homo sapiens],3GXF_A Crystal structure of acid-beta-glucosidase with isofagomine at neutral pH [Homo sapiens],3GXF_B Crystal structure of acid-beta-glucosidase with isofagomine at neutral pH [Homo sapiens],3GXF_C Crystal structure of acid-beta-glucosidase with isofagomine at neutral pH [Homo sapiens],3GXF_D Crystal structure of acid-beta-glucosidase with isofagomine at neutral pH [Homo sapiens],3GXI_A Crystal structure of acid-beta-glucosidase at pH 5.5 [Homo sapiens],3GXI_B Crystal structure of acid-beta-glucosidase at pH 5.5 [Homo sapiens],3GXI_C Crystal structure of acid-beta-glucosidase at pH 5.5 [Homo sapiens],3GXI_D Crystal structure of acid-beta-glucosidase at pH 5.5 [Homo sapiens],3GXM_A Crystal structure of acid-beta-glucosidase at pH 4.5, phosphate crystallization condition [Homo sapiens],3GXM_B Crystal structure of acid-beta-glucosidase at pH 4.5, phosphate crystallization condition [Homo sapiens],3GXM_C Crystal structure of acid-beta-glucosidase at pH 4.5, phosphate crystallization condition [Homo sapiens],3GXM_D Crystal structure of acid-beta-glucosidase at pH 4.5, phosphate crystallization condition [Homo sapiens],3RIK_A The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIK_B The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIK_C The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIK_D The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIL_A The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIL_B The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIL_C The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],3RIL_D The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease [Homo sapiens],6MOZ_A Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor [Homo sapiens],6MOZ_B Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor [Homo sapiens],6Q1N_A Glucocerebrosidase in complex with pharmacological chaperone IMX8 [Homo sapiens],6Q1N_B Glucocerebrosidase in complex with pharmacological chaperone IMX8 [Homo sapiens],6Q1P_A Glucocerebrosidase in complex with pharmacological chaperone norIMX8 [Homo sapiens],6Q1P_B Glucocerebrosidase in complex with pharmacological chaperone norIMX8 [Homo sapiens],6Q6K_A Crystal structure of recombinant human beta-glucocerebrosidase in complex with cyclophellitol activity based probe with Cy5 tag (ME569) [Homo sapiens],6Q6K_B Crystal structure of recombinant human beta-glucocerebrosidase in complex with cyclophellitol activity based probe with Cy5 tag (ME569) [Homo sapiens],6Q6L_A Crystal structure of recombinant human beta-glucocerebrosidase in complex with adamantyl-cyclophellitol inhibitor (ME656) [Homo sapiens],6Q6L_B Crystal structure of recombinant human beta-glucocerebrosidase in complex with adamantyl-cyclophellitol inhibitor (ME656) [Homo sapiens],6Q6N_A Crystal structure of recombinant human beta-glucocerebrosidase in complex with biphenyl-cyclophellitol inhibitor (ME655) [Homo sapiens],6Q6N_B Crystal structure of recombinant human beta-glucocerebrosidase in complex with biphenyl-cyclophellitol inhibitor (ME655) [Homo sapiens],6TJJ_AAA Chain AAA, Glucosylceramidase [Homo sapiens],6TJJ_BBB Chain BBB, Glucosylceramidase [Homo sapiens],6YTP_AAA Chain AAA, Glucosylceramidase [Homo sapiens],6YTP_BBB Chain BBB, Glucosylceramidase [Homo sapiens],6YUT_AAA Chain AAA, Glucosylceramidase [Homo sapiens],6YUT_BBB Chain BBB, Glucosylceramidase [Homo sapiens],6YV3_AAA Chain AAA, Glucosylceramidase [Homo sapiens],6YV3_BBB Chain BBB, Glucosylceramidase [Homo sapiens],6Z39_AAA Chain AAA, Glucosylceramidase [Homo sapiens],6Z39_BBB Chain BBB, Glucosylceramidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4WBR2 | 3.95e-45 | 72 | 496 | 54 | 483 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
| Q7M4T0 | 9.07e-45 | 50 | 496 | 23 | 473 | Endo-1,6-beta-D-glucanase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=neg-1 PE=1 SV=2 |
| Q8J0I9 | 2.52e-41 | 74 | 496 | 61 | 485 | Endo-1,6-beta-D-glucanase BGN16.3 OS=Trichoderma harzianum OX=5544 PE=1 SV=1 |
| O16580 | 9.36e-39 | 46 | 495 | 55 | 515 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
| O16581 | 1.47e-37 | 44 | 468 | 53 | 481 | Putative glucosylceramidase 2 OS=Caenorhabditis elegans OX=6239 GN=gba-2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
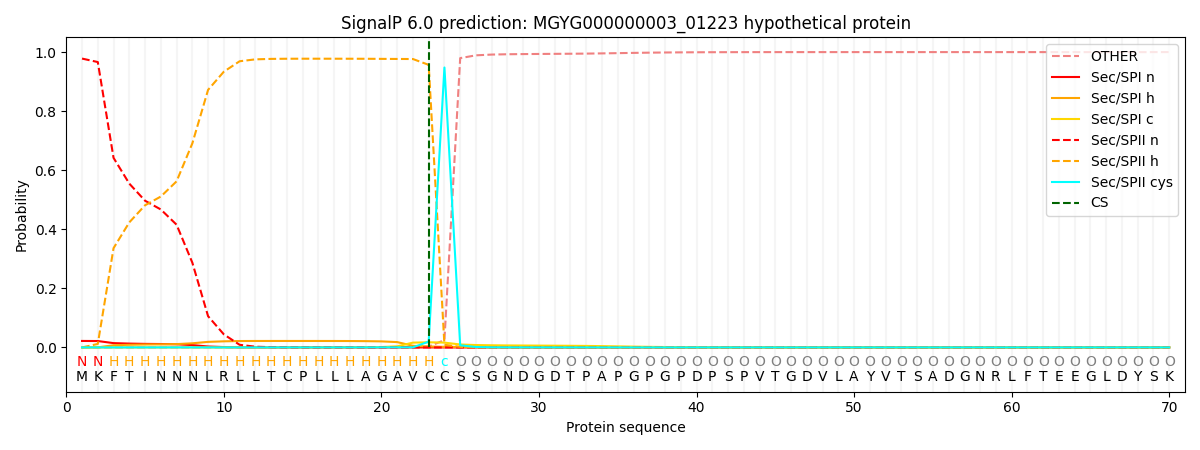
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000060 | 0.021350 | 0.978575 | 0.000009 | 0.000012 | 0.000010 |
