You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000003_02605
You are here: Home > Sequence: MGYG000000003_02605
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes shahii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes shahii | |||||||||||
| CAZyme ID | MGYG000000003_02605 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | Alpha-1,3-galactosidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13064; End: 14929 Strand: + | |||||||||||
Full Sequence Download help
| MFGMSIRKFI ILSALIVSGC ISHPETIDID FDSGTEDYTP LVRKILAEHP AGEVTIRFGA | 60 |
| GTFDFYPEQA AGSYLCVSNN DNGYKRCAFL LEEMRRVRIE GTGEKTQLRF HGAIVPFRVA | 120 |
| RCEQIVFEAF TIDYDASFIF EGLVVGNDPR THSITLRPLD PERFEIRSGE PWFTGYDWAS | 180 |
| PFGENILFDP GTRSPYYQAE QYEHDQKKTL QAEWIGDSLV RLSGYSSREL PPVGSVYTDK | 240 |
| GPHSTNRRYP GFSFYKSAGV EVRNVTLHDS GGMALIAENC RNVVCSQYRV EVPPQSGRMV | 300 |
| SASADATHFV GCSGKIVLRA CRFESMLDDA TNIHGVYMTV VDRLSGNRFG ASFGHFQQEG | 360 |
| FDFAEQGDSL VFIDRADLGV LGCGRVEEVN HVNENYYIIR TGFDLSTIPD SVHIAVGNRA | 420 |
| ADADVEISEC TVRYNRARSF LLSTPGDVCV ENSDLSSMMA GIRICGDANY WFESGRTRNV | 480 |
| VIRNNRFGTM ATGGCSPQAV LQIDPVISHN ARSGGTPYHG CIRFEGNLVE SFDNQLIYAL | 540 |
| SVDSLVISRN RFVDSRRFEP RFAGLSVIDA QHCRSVTVRD NDFSGWKENS TISLVDCSEH | 600 |
| CLEGEEMPRM VENPNPYFYE N | 621 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 34 | 581 | 2.2e-130 | 0.9762773722627737 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFL77292.1 | 0.0 | 1 | 621 | 1 | 621 |
| QDH53926.1 | 6.06e-205 | 16 | 621 | 18 | 620 |
| EDM26187.1 | 2.95e-107 | 36 | 594 | 36 | 594 |
| QDV61916.1 | 4.14e-103 | 34 | 598 | 32 | 591 |
| AFL78386.1 | 1.66e-98 | 35 | 595 | 37 | 591 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 3.05e-59 | 22 | 591 | 23 | 606 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 1.54e-58 | 22 | 591 | 23 | 606 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1KD83 | 1.46e-62 | 36 | 594 | 46 | 593 | Alpha-1,3-galactosidase A OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=glaA PE=3 SV=1 |
| Q89ZX0 | 1.17e-60 | 40 | 585 | 45 | 586 | Alpha-1,3-galactosidase B OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaB PE=3 SV=1 |
| A6LFT2 | 7.33e-58 | 54 | 562 | 62 | 571 | Alpha-1,3-galactosidase B OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=glaB PE=3 SV=1 |
| A6KWM0 | 1.87e-57 | 44 | 580 | 49 | 592 | Alpha-1,3-galactosidase B OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=glaB1 PE=3 SV=1 |
| Q5LGZ8 | 1.84e-55 | 39 | 560 | 43 | 575 | Alpha-1,3-galactosidase B OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
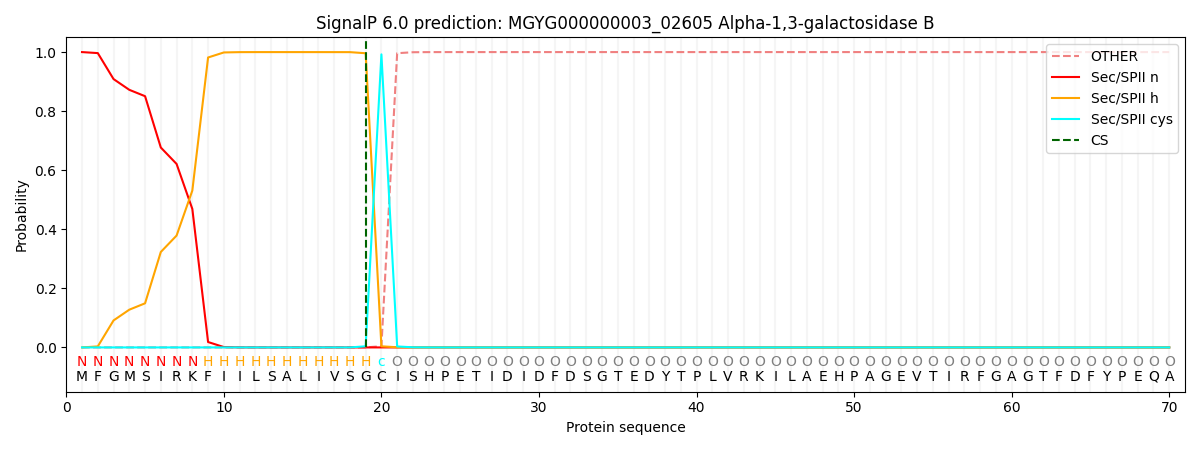
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000002 | 1.000053 | 0.000000 | 0.000000 | 0.000000 |
