You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000006_00015
You are here: Home > Sequence: MGYG000000006_00015
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
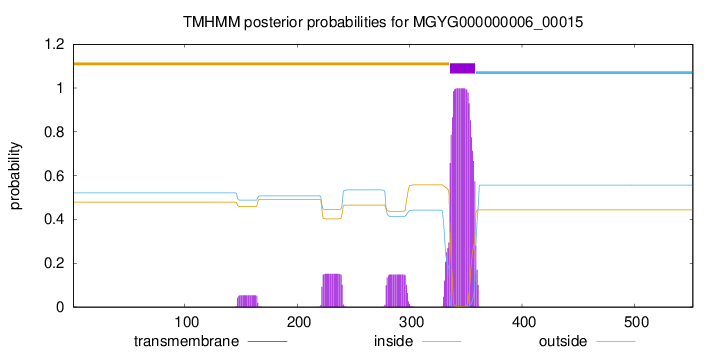
TMHMM annotations
Basic Information help
| Species | Staphylococcus xylosus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus xylosus | |||||||||||
| CAZyme ID | MGYG000000006_00015 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12629; End: 14287 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033598 | elast_bind_EbpS | 1.03e-23 | 5 | 551 | 1 | 466 | elastin-binding protein EbpS. The elastin-binding protein EbpS is an adhesin described in Staphylococcus aureus, with orthologs found in many additional staphylococcal species. EbpS is a membrane protein that lacks an N-terminal signal peptide region, has extensive regions low-complexity sequence rich in Asn and Gln, and has a C-terminal LysM domain. |
| pfam01476 | LysM | 5.73e-05 | 505 | 551 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| COG5271 | MDN1 | 5.20e-04 | 17 | 278 | 3825 | 4089 | Midasin, AAA ATPase with vWA domain, involved in ribosome maturation [Translation, ribosomal structure and biogenesis]. |
| COG5271 | MDN1 | 0.001 | 13 | 217 | 3929 | 4124 | Midasin, AAA ATPase with vWA domain, involved in ribosome maturation [Translation, ribosomal structure and biogenesis]. |
| PRK12678 | PRK12678 | 0.003 | 14 | 213 | 56 | 254 | transcription termination factor Rho; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDW89145.1 | 1.22e-121 | 1 | 552 | 1 | 552 |
| ARD74584.1 | 1.99e-66 | 1 | 552 | 1 | 542 |
| QRA15459.1 | 2.00e-40 | 1 | 139 | 1 | 143 |
| QKV11442.1 | 2.55e-28 | 1 | 169 | 2 | 172 |
| QKQ10341.1 | 1.08e-22 | 1 | 169 | 2 | 172 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q49XT4 | 2.19e-20 | 3 | 169 | 1 | 169 | Probable elastin-binding protein EbpS OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=ebpS PE=4 SV=1 |
| Q2YY76 | 3.51e-13 | 507 | 551 | 438 | 483 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain bovine RF122 / ET3-1) OX=273036 GN=ebpS PE=3 SV=3 |
| Q6GGT1 | 3.54e-13 | 507 | 551 | 441 | 486 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=ebpS PE=3 SV=3 |
| Q6G983 | 3.54e-13 | 507 | 551 | 441 | 486 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=ebpS PE=3 SV=3 |
| Q5HFU2 | 3.54e-13 | 507 | 551 | 441 | 486 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain COL) OX=93062 GN=ebpS PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
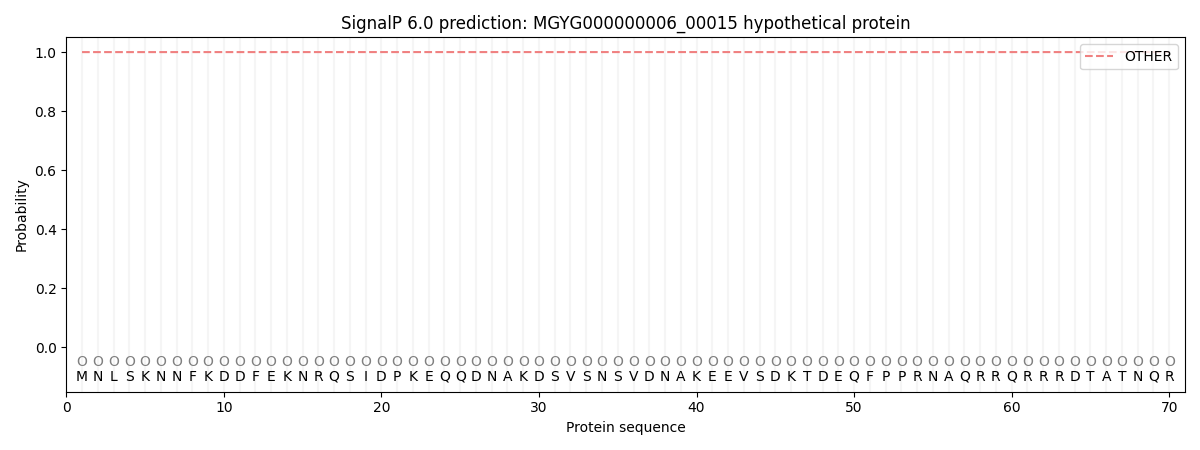
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |