You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000009_01920
You are here: Home > Sequence: MGYG000000009_01920
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ligilactobacillus murinus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Ligilactobacillus; Ligilactobacillus murinus | |||||||||||
| CAZyme ID | MGYG000000009_01920 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 595; End: 1941 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 56 | 239 | 1.5e-32 | 0.9887005649717514 |
| CBM50 | 354 | 396 | 1.8e-16 | 0.95 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06415 | GH25_Cpl1-like | 4.58e-63 | 54 | 246 | 3 | 196 | Cpl-1 lysin (also known as Cpl-9 lysozyme / muramidase) is a bacterial cell wall endolysin encoded by the pneumococcal bacteriophage Cp-1, which cleaves the glycosidic N-acetylmuramoyl-(beta1,4)-N-acetylglucosamine bonds of the pneumococcal glycan chain, thus acting as an enzymatic antimicrobial agent (an enzybiotic) against streptococcal infections. Cpl-1 belongs to the CP family of lysozymes (CPL lysozymes) which includes the Cpl-7 lysin. Cpl-1 has a glycosyl hydrolase family 25 (GH25) catalytic domain with an irregular (beta/alpha)5-beta3 barrel and a C-terminal cell wall-anchoring module formed by six similar choline-binding repeats (ChBr's). The ChBr's facilitate the anchoring of Cpl-1 to the choline-containing teichoic acid of the pneumococcal cell wall. Other members of this domain family have an N-terminal CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain similar to that of the firmicute CHAP lysins and associated with endopeptidase activity. The Cpl-7 lysin is also included here as is LysB of Lactococcus phage, and the Mur lysin of Lactobacillus phage. |
| pfam01183 | Glyco_hydro_25 | 4.76e-23 | 56 | 239 | 2 | 180 | Glycosyl hydrolases family 25. |
| smart00641 | Glyco_25 | 1.72e-22 | 137 | 249 | 1 | 106 | Glycosyl hydrolases family 25. |
| PRK10783 | mltD | 1.10e-16 | 332 | 439 | 326 | 438 | membrane-bound lytic murein transglycosylase D; Provisional |
| PRK06347 | PRK06347 | 5.72e-16 | 307 | 446 | 411 | 589 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA90198.1 | 3.68e-275 | 3 | 448 | 2 | 429 |
| QCQ04151.1 | 1.58e-249 | 1 | 448 | 1 | 451 |
| ASD50641.1 | 4.54e-221 | 4 | 395 | 2 | 393 |
| AWZ37749.1 | 9.01e-170 | 1 | 448 | 1 | 419 |
| AWZ41262.1 | 9.01e-170 | 1 | 448 | 1 | 419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1H09_A | 8.35e-18 | 110 | 246 | 58 | 187 | ChainA, LYSOZYME [Streptococcus phage Cp1] |
| 1OBA_A | 8.44e-18 | 110 | 246 | 59 | 188 | ChainA, Lysozyme [Streptococcus phage Cp1] |
| 2IXU_A | 8.44e-18 | 110 | 246 | 59 | 188 | ChainA, LYSOZYME [Streptococcus phage Cp1] |
| 2IXV_A | 2.07e-17 | 110 | 246 | 59 | 188 | ChainA, LYSOZYME [Streptococcus phage Cp1],2J8F_A Chain A, LYSOZYME [Streptococcus phage Cp1],2J8G_A Chain A, LYSOZYME [Streptococcus phage Cp1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8HA43 | 1.03e-33 | 34 | 246 | 135 | 343 | D-alanyl-L-alanine endopeptidase OS=Streptococcus phage B30 OX=209152 PE=1 SV=2 |
| Q49UX4 | 2.02e-20 | 353 | 447 | 87 | 192 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| Q4L3C1 | 3.84e-20 | 350 | 447 | 26 | 130 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=sle1 PE=3 SV=1 |
| P19386 | 6.78e-19 | 110 | 246 | 59 | 188 | Lysozyme OS=Streptococcus phage Cp-9 OX=10749 GN=CPL9 PE=3 SV=1 |
| P19385 | 1.29e-18 | 50 | 251 | 4 | 193 | Lysozyme OS=Streptococcus phage Cp-7 OX=10748 GN=CPL7 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
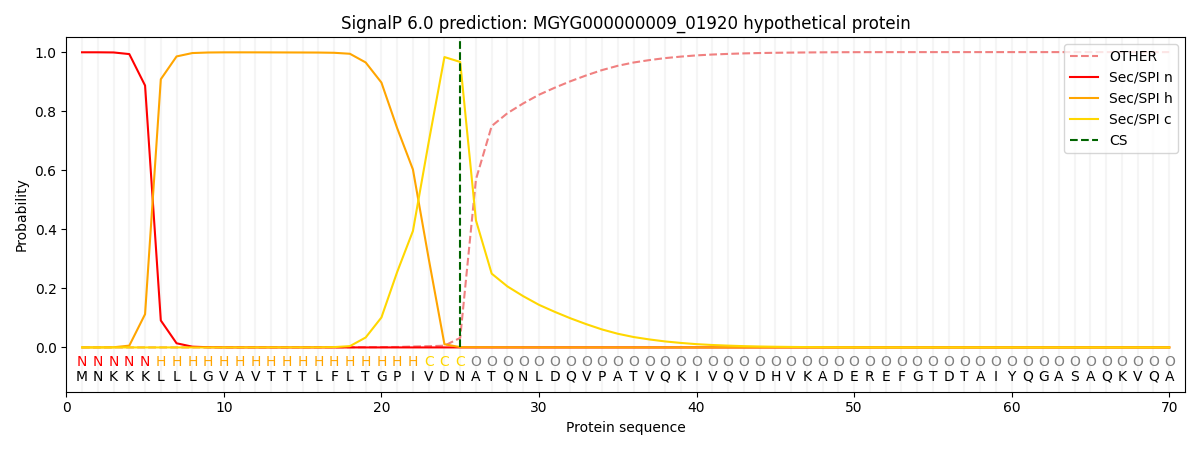
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000445 | 0.998071 | 0.000928 | 0.000201 | 0.000168 | 0.000150 |
