You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_00158
You are here: Home > Sequence: MGYG000000012_00158
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_00158 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Peptidoglycan endopeptidase LytF | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 126300; End: 127766 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK06347 | PRK06347 | 3.93e-50 | 35 | 349 | 271 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 4.96e-45 | 24 | 285 | 328 | 592 | 1,4-beta-N-acetylmuramoylhydrolase. |
| pfam00877 | NLPC_P60 | 4.34e-40 | 385 | 486 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK06347 | PRK06347 | 2.76e-36 | 174 | 350 | 332 | 524 | 1,4-beta-N-acetylmuramoylhydrolase. |
| COG0791 | Spr | 7.22e-34 | 376 | 485 | 78 | 197 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFI27572.1 | 4.10e-290 | 1 | 488 | 1 | 488 |
| QRZ93882.1 | 4.10e-290 | 1 | 488 | 1 | 488 |
| AUS13182.1 | 2.37e-289 | 1 | 488 | 1 | 488 |
| ASB60224.1 | 6.77e-289 | 1 | 488 | 1 | 488 |
| AUZ37869.1 | 1.87e-288 | 1 | 488 | 1 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CFL_A | 2.39e-19 | 374 | 486 | 15 | 136 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4XCM_A | 1.02e-16 | 241 | 486 | 5 | 229 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 2K1G_A | 1.00e-15 | 385 | 486 | 18 | 122 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 6B8C_A | 5.19e-13 | 376 | 466 | 31 | 123 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 3PBI_A | 3.51e-09 | 313 | 465 | 23 | 190 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O07532 | 6.82e-262 | 1 | 488 | 1 | 488 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| O31852 | 4.37e-140 | 1 | 488 | 1 | 414 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| P54421 | 3.61e-90 | 176 | 487 | 29 | 334 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| P39046 | 4.79e-35 | 24 | 349 | 333 | 664 | Muramidase-2 OS=Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R) OX=768486 GN=EHR_05900 PE=1 SV=1 |
| Q6GJK9 | 7.42e-35 | 174 | 475 | 28 | 311 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
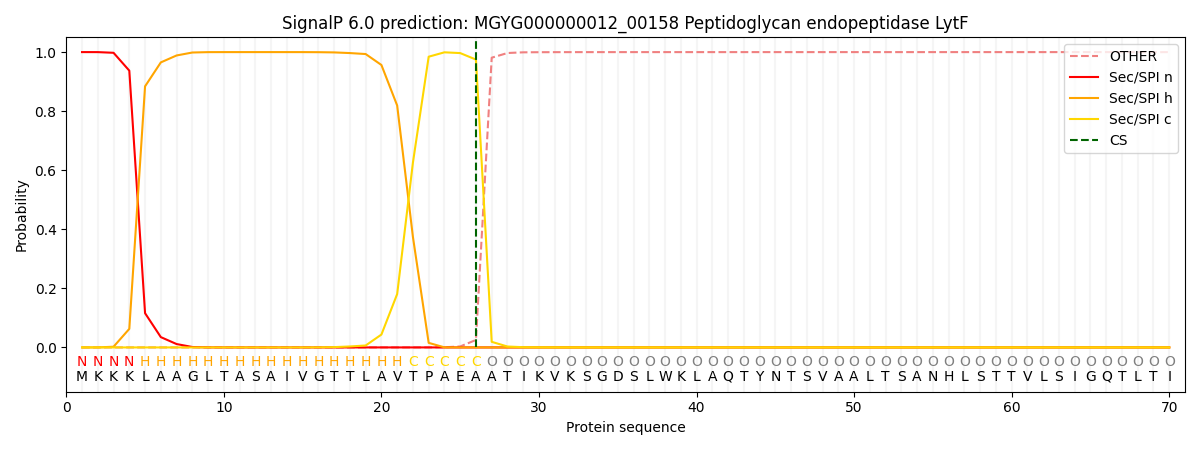
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998815 | 0.000188 | 0.000243 | 0.000220 | 0.000187 |
