You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_00998
You are here: Home > Sequence: MGYG000000012_00998
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_00998 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1006095; End: 1006793 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 40 | 221 | 5.9e-77 | 0.9943502824858758 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 1.10e-87 | 40 | 219 | 1 | 175 | Glycosyl hydrolases family 11. |
| PRK06215 | PRK06215 | 0.006 | 1 | 185 | 8 | 192 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUS13960.1 | 3.56e-169 | 1 | 232 | 1 | 232 |
| ASB61035.1 | 1.45e-168 | 1 | 232 | 1 | 232 |
| QRZ94754.1 | 5.91e-168 | 1 | 232 | 1 | 232 |
| QJC96409.1 | 1.10e-165 | 1 | 230 | 1 | 230 |
| AEP86749.1 | 2.58e-165 | 1 | 232 | 1 | 232 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1IGO_A | 2.01e-145 | 28 | 230 | 1 | 203 | Family11 xylanase [Bacillus subtilis],1IGO_B Family 11 xylanase [Bacillus subtilis] |
| 2DCJ_A | 4.46e-111 | 1 | 231 | 1 | 231 | Atwo-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCJ_B A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCK_A A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 6KKA_A | 2.82e-108 | 31 | 231 | 2 | 204 | XylanaseJ mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],6KKA_B Xylanase J mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 6KJL_A | 4.14e-108 | 31 | 231 | 3 | 205 | XylanaseJ from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1],6KJL_B Xylanase J from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1] |
| 2F6B_A | 1.03e-106 | 29 | 229 | 1 | 203 | Structuraland active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus],2F6B_B Structural and active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P00694 | 2.39e-126 | 1 | 228 | 1 | 227 | Endo-1,4-beta-xylanase A OS=Bacillus pumilus OX=1408 GN=xynA PE=1 SV=2 |
| P17137 | 4.55e-106 | 19 | 228 | 49 | 260 | Endo-1,4-beta-xylanase OS=Clostridium saccharobutylicum OX=169679 GN=xynB PE=3 SV=1 |
| Q8GJ44 | 1.54e-99 | 11 | 228 | 14 | 232 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P83513 | 2.80e-98 | 15 | 231 | 2 | 219 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
| P33558 | 3.60e-96 | 11 | 228 | 14 | 233 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
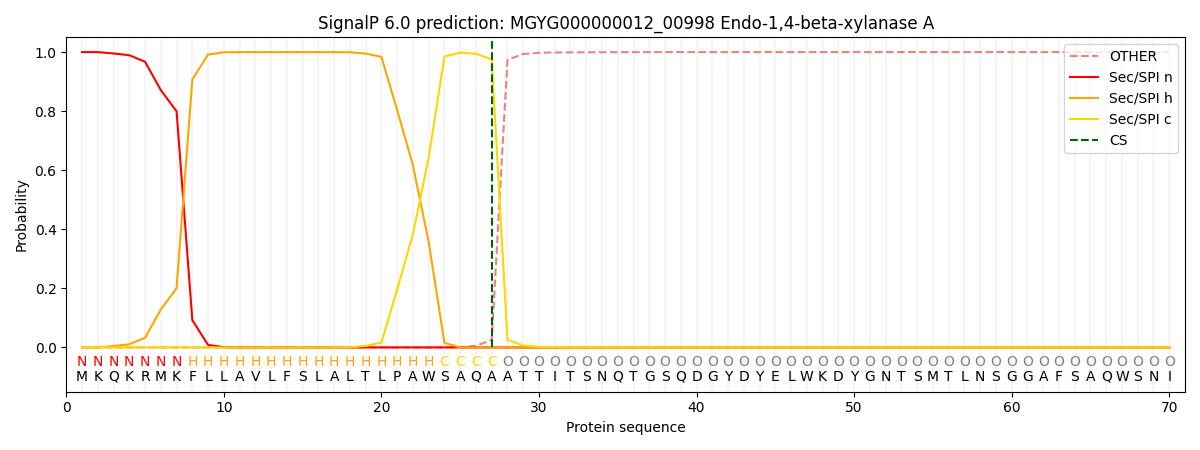
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.998970 | 0.000184 | 0.000228 | 0.000177 | 0.000155 |
