You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_02573
You are here: Home > Sequence: MGYG000000012_02573
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_02573 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | Levanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 413998; End: 416031 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH32 | 39 | 349 | 4.3e-109 | 0.9965870307167235 |
| CBM66 | 520 | 673 | 7.1e-55 | 0.9935483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1621 | SacC | 0.0 | 7 | 511 | 1 | 486 | Sucrose-6-phosphate hydrolase SacC, GH32 family [Carbohydrate transport and metabolism]. |
| cd18622 | GH32_Inu-like | 2.03e-177 | 44 | 336 | 1 | 289 | glycoside hydrolase family 32 protein such as Aspergillus ficuum endo-inulinase (Inu2). This subfamily of glycosyl hydrolase family GH32 includes endo-inulinase (inu2, EC 3.2.1.7), exo-inulinase (Inu1, EC 3.2.1.80), invertase (EC 3.2.1.26), and levan fructotransferase (LftA, EC 4.2.2.16), among others. These enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| smart00640 | Glyco_32 | 5.84e-173 | 39 | 473 | 1 | 437 | Glycosyl hydrolases family 32. |
| pfam00251 | Glyco_hydro_32N | 6.13e-138 | 39 | 349 | 1 | 308 | Glycosyl hydrolases family 32 N-terminal domain. This domain corresponds to the N-terminal domain of glycosyl hydrolase family 32 which forms a five bladed beta propeller structure. |
| cd08996 | GH32_FFase | 1.23e-100 | 45 | 336 | 1 | 281 | Glycosyl hydrolase family 32, beta-fructosidases. Glycosyl hydrolase family GH32 cleaves sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRZ91563.1 | 0.0 | 1 | 677 | 1 | 677 |
| AFI29216.1 | 0.0 | 1 | 677 | 9 | 685 |
| ASB61793.1 | 0.0 | 1 | 677 | 1 | 677 |
| AUS10781.1 | 0.0 | 1 | 677 | 1 | 677 |
| AUZ39607.1 | 0.0 | 1 | 677 | 1 | 677 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1Y4W_A | 1.78e-122 | 30 | 513 | 3 | 517 | Crystalstructure of exo-inulinase from Aspergillus awamori in spacegroup P21 [Aspergillus awamori],1Y9G_A Crystal structure of exo-inulinase from Aspergillus awamori complexed with fructose [Aspergillus awamori],1Y9M_A Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121 [Aspergillus awamori] |
| 4B1L_A | 1.74e-102 | 515 | 677 | 3 | 165 | CarbohydrateBinding Module Cbm66 From Bacillus Subtilis [Bacillus subtilis] |
| 4AZZ_A | 2.22e-102 | 515 | 677 | 2 | 164 | Carbohydratebinding module CBM66 from Bacillus subtilis [Bacillus subtilis],4AZZ_B Carbohydrate binding module CBM66 from Bacillus subtilis [Bacillus subtilis] |
| 3RWK_X | 2.73e-102 | 33 | 506 | 27 | 510 | Firstcrystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. [Aspergillus ficuum],3SC7_X First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. [Aspergillus ficuum] |
| 4B1M_A | 3.49e-102 | 515 | 677 | 23 | 185 | CarbohydrateBinding Module Cbm66 From Bacillus Subtilis [Bacillus subtilis],4B1M_B Carbohydrate Binding Module Cbm66 From Bacillus Subtilis [Bacillus subtilis],4B1M_C Carbohydrate Binding Module Cbm66 From Bacillus Subtilis [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P05656 | 0.0 | 1 | 677 | 1 | 677 | Levanase OS=Bacillus subtilis (strain 168) OX=224308 GN=sacC PE=1 SV=1 |
| O31411 | 7.33e-143 | 27 | 513 | 390 | 879 | Levanase (Fragment) OS=Bacillus sp. (strain L7) OX=62626 PE=1 SV=2 |
| Q76HP6 | 5.09e-124 | 16 | 513 | 8 | 536 | Extracellular exo-inulinase inuE OS=Aspergillus niger OX=5061 GN=inuE PE=1 SV=1 |
| E1ABX2 | 5.09e-124 | 16 | 513 | 8 | 536 | Extracellular exo-inulinase inuE OS=Aspergillus ficuum OX=5058 GN=exoI PE=1 SV=1 |
| A2R0E0 | 4.00e-123 | 16 | 513 | 8 | 536 | Extracellular exo-inulinase inuE OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=inuE PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
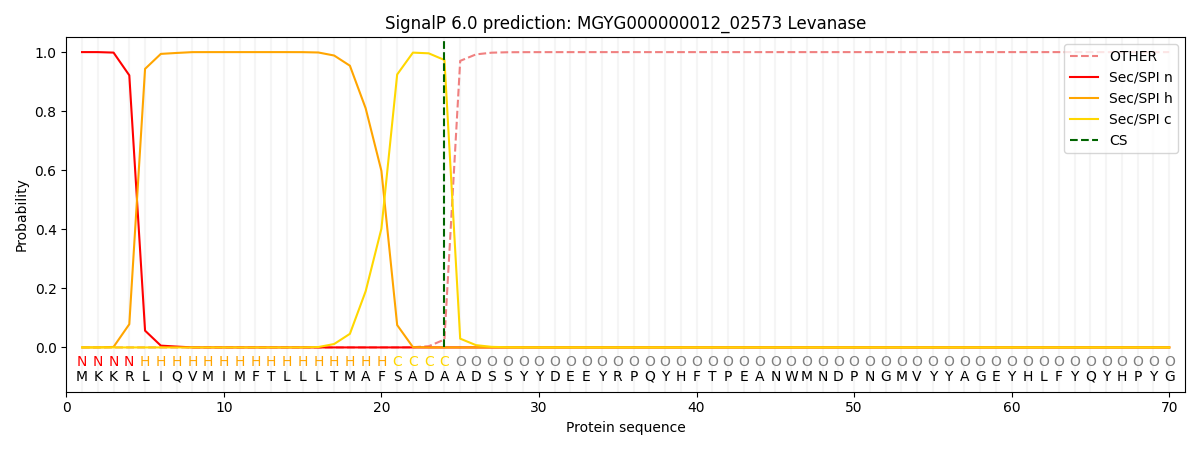
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000274 | 0.998857 | 0.000247 | 0.000225 | 0.000197 | 0.000184 |
