You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_03213
You are here: Home > Sequence: MGYG000000012_03213
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_03213 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Pectin lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 972939; End: 973976 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 83 | 274 | 1.6e-91 | 0.9948717948717949 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 2.89e-125 | 4 | 336 | 1 | 341 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 6.28e-58 | 92 | 277 | 7 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 2.32e-44 | 93 | 273 | 26 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRZ90970.1 | 4.29e-254 | 1 | 345 | 1 | 345 |
| AUS14071.1 | 4.29e-254 | 1 | 345 | 1 | 345 |
| AFI28572.1 | 2.04e-252 | 1 | 345 | 1 | 345 |
| ASB61140.1 | 4.80e-251 | 1 | 345 | 1 | 345 |
| AUZ38955.1 | 1.38e-250 | 1 | 345 | 1 | 345 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1PCL_A | 2.02e-16 | 53 | 322 | 17 | 337 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
| 1OOC_A | 7.55e-11 | 54 | 273 | 24 | 287 | ChainA, Pectate lyase A [Dickeya chrysanthemi],1OOC_B Chain B, Pectate lyase A [Dickeya chrysanthemi],1PE9_A Chain A, Pectate lyase A [Dickeya chrysanthemi],1PE9_B Chain B, Pectate lyase A [Dickeya chrysanthemi] |
| 1JRG_A | 1.01e-10 | 54 | 273 | 24 | 287 | ChainA, Pectate lyase [Dickeya chrysanthemi],1JRG_B Chain B, Pectate lyase [Dickeya chrysanthemi],1JTA_A Chain A, pectate lyase A [Dickeya chrysanthemi] |
| 1QCX_A | 1.03e-09 | 53 | 273 | 13 | 276 | PectinLyase B [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34819 | 2.53e-247 | 1 | 345 | 1 | 345 | Pectin lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pelB PE=3 SV=1 |
| P94449 | 3.59e-247 | 1 | 345 | 1 | 345 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| P27027 | 5.09e-105 | 26 | 344 | 1 | 311 | Pectin lyase OS=Pseudomonas marginalis OX=298 GN=pnl PE=1 SV=2 |
| P24112 | 6.13e-99 | 28 | 343 | 3 | 312 | Pectin lyase OS=Pectobacterium carotovorum OX=554 GN=pnl PE=1 SV=1 |
| Q0CBV0 | 1.42e-23 | 53 | 253 | 45 | 236 | Probable pectate lyase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
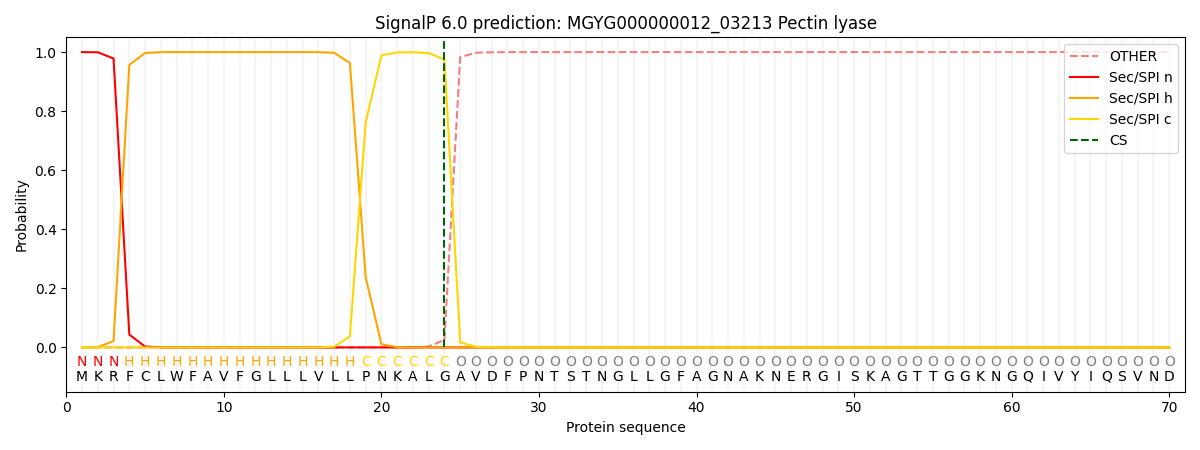
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000275 | 0.999055 | 0.000192 | 0.000164 | 0.000157 | 0.000157 |