You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_03331
You are here: Home > Sequence: MGYG000000012_03331
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_03331 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 103270; End: 107793 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00833 | PKS | 0.0 | 8 | 428 | 2 | 421 | polyketide synthases (PKSs) polymerize simple fatty acids into a large variety of different products, called polyketides, by successive decarboxylating Claisen condensations. PKSs can be divided into 2 groups, modular type I PKSs consisting of one or more large multifunctional proteins and iterative type II PKSs, complexes of several monofunctional subunits. |
| COG3321 | PksD | 0.0 | 9 | 871 | 6 | 870 | Acyl transferase domain in polyketide synthase (PKS) enzymes [Secondary metabolites biosynthesis, transport and catabolism]. |
| smart00825 | PKS_KS | 1.97e-137 | 9 | 430 | 1 | 298 | Beta-ketoacyl synthase. The structure of beta-ketoacyl synthase is similar to that of the thiolase family and also chalcone synthase. The active site of beta-ketoacyl synthase is located between the N and C-terminal domains. |
| TIGR02813 | omega_3_PfaA | 2.02e-95 | 9 | 827 | 9 | 901 | polyketide-type polyunsaturated fatty acid synthase PfaA. Members of the seed for this alignment are involved in omega-3 polyunsaturated fatty acid biosynthesis, such as the protein PfaA from the eicosapentaenoic acid biosynthesis operon in Photobacterium profundum strain SS9. PfaA is encoded together with PfaB, PfaC, and PfaD, and the functions of the individual polypeptides have not yet been described. More distant homologs of PfaA, also included with the reach of this model, appear to be involved in polyketide-like biosynthetic mechanisms of polyunsaturated fatty acid biosynthesis, an alternative to the more familiar iterated mechanism of chain extension and desaturation, and in most cases are encoded near genes for homologs of PfaB, PfaC, and/or PfaD. |
| smart00827 | PKS_AT | 1.34e-94 | 522 | 802 | 1 | 280 | Acyl transferase domain in polyketide synthase (PKS) enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAY30132.1 | 2.75e-239 | 8 | 871 | 1215 | 2094 |
| BAZ00088.1 | 2.12e-237 | 8 | 871 | 1213 | 2092 |
| BAZ75991.1 | 2.12e-237 | 8 | 871 | 1213 | 2092 |
| BAY90071.1 | 2.74e-237 | 8 | 871 | 1212 | 2091 |
| AFY93865.1 | 2.84e-221 | 3 | 871 | 957 | 1860 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MZ0_A | 1.94e-173 | 9 | 871 | 41 | 934 | ChainA, CurL [Moorena producens 3L],4MZ0_B Chain B, CurL [Moorena producens 3L] |
| 7M7J_A | 2.72e-156 | 6 | 1474 | 32 | 1488 | ChainA, EryAI [Saccharopolyspora erythraea],7M7J_B Chain B, EryAI [Saccharopolyspora erythraea] |
| 7M7I_A | 7.15e-156 | 6 | 1467 | 32 | 1481 | ChainA, EryAI [Saccharopolyspora erythraea],7M7I_B Chain B, EryAI [Saccharopolyspora erythraea] |
| 7M7F_A | 1.26e-154 | 6 | 1467 | 32 | 1481 | ChainA, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea],7M7F_B Chain B, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea],7M7G_A Chain A, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea],7M7G_B Chain B, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea],7M7H_A Chain A, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea],7M7H_B Chain B, EryAI,6-deoxyerythronolide-B synthase EryA3, modules 5 and 6 chimera [Saccharopolyspora erythraea] |
| 4WKY_A | 3.92e-149 | 5 | 518 | 5 | 514 | Streptomcyesalbus JA3453 oxazolomycin ketosynthase domain OzmN KS2 [Streptomyces albus],4WKY_B Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 [Streptomyces albus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7TXL6 | 3.69e-197 | 9 | 869 | 8 | 891 | Phenolphthiocerol/phthiocerol polyketide synthase subunit E OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=ppsE PE=1 SV=1 |
| P9WQE0 | 3.69e-197 | 9 | 869 | 8 | 891 | Phenolphthiocerol/phthiocerol polyketide synthase subunit E OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=ppsE PE=3 SV=1 |
| P9WQE1 | 3.69e-197 | 9 | 869 | 8 | 891 | Phenolphthiocerol/phthiocerol polyketide synthase subunit E OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ppsE PE=1 SV=1 |
| D4AU31 | 8.72e-159 | 8 | 1386 | 626 | 1966 | PKS-NRPS hybrid synthetase swnK OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=swnK PE=3 SV=1 |
| Q03131 | 3.40e-152 | 6 | 1477 | 504 | 1963 | 6-deoxyerythronolide-B synthase EryA1, modules 1 and 2 OS=Saccharopolyspora erythraea OX=1836 GN=eryA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
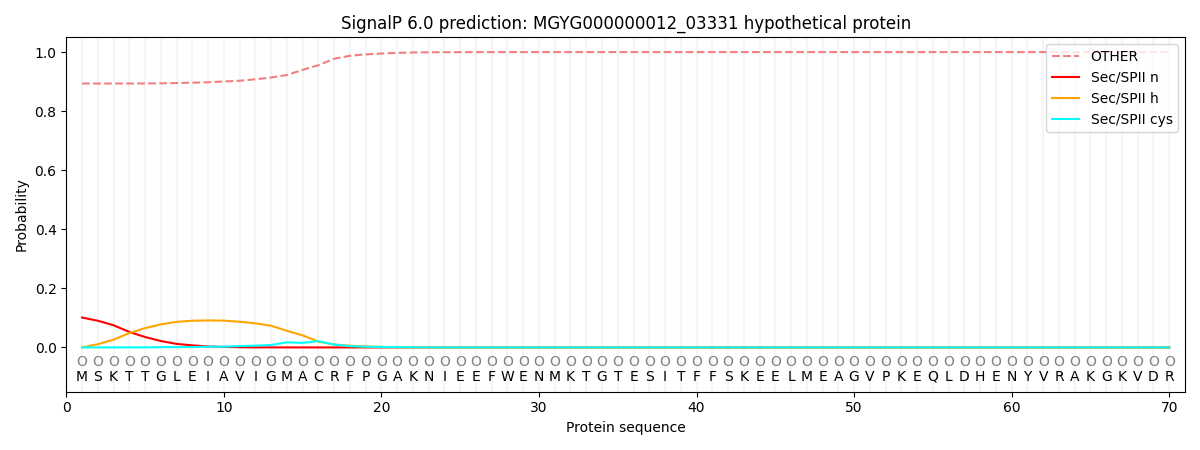
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.894107 | 0.004505 | 0.101248 | 0.000027 | 0.000013 | 0.000127 |
