You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000012_03445
You are here: Home > Sequence: MGYG000000012_03445
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
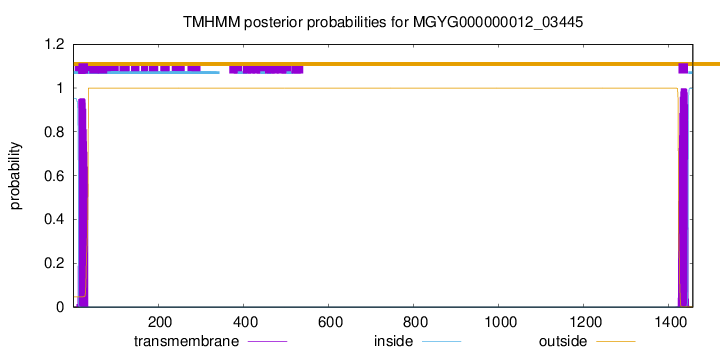
TMHMM annotations
Basic Information help
| Species | Bacillus subtilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus subtilis | |||||||||||
| CAZyme ID | MGYG000000012_03445 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 239242; End: 243615 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 0.0 | 5 | 1194 | 1 | 1163 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| PRK11907 | PRK11907 | 0.0 | 29 | 662 | 96 | 734 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| PRK09418 | PRK09418 | 0.0 | 27 | 665 | 22 | 673 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| TIGR01390 | CycNucDiestase | 0.0 | 43 | 622 | 1 | 590 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase. 2',3'-cyclic-nucleotide 2'-phosphodiesterase is a bifunctional enzyme localized to the periplasm of Gram-negative bacteria. 2',3'-cyclic-nucleotide 2'-phosphodiesters are intermediates formed during the hydrolysis of RNA by the ribonuclease I, which is also found to the periplasm, and other enzymes of the RNAse T2 family. Bacteria are unable to transport 2',3'-cyclic-nucleotides into the cytoplasm. 2',3'-cyclic-nucleotide 2'-phosphodiesterase contains 2 active sites which catalyze the reactions that convert the 2',3'-cyclic-nucleotide into a 3'-nucleotide, which is then converted into nucleic acid and phosphate. Both final products can be transported into the cytoplasm. Thus, it has been suggested that 2',3'-cyclic-nucleotide 2'-phosphodiesterase has a 'scavenging' function. Experimental evidence indicates that 2',3'-cyclic-nucleotide 2'-phosphodiesterase enables Yersinia enterocolitica O:8 to grow on 2'3'-cAMP as a sole source of carbon and energy (). [Purines, pyrimidines, nucleosides, and nucleotides, Other] |
| PRK09420 | cpdB | 0.0 | 39 | 651 | 20 | 645 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLG37215.1 | 7.19e-229 | 32 | 661 | 25 | 671 |
| APO43595.1 | 2.22e-228 | 32 | 661 | 23 | 669 |
| QZN77538.1 | 1.31e-227 | 32 | 661 | 25 | 671 |
| QOS76560.1 | 2.17e-226 | 32 | 661 | 25 | 671 |
| QKS58250.1 | 4.40e-225 | 32 | 652 | 25 | 662 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GVE_A | 2.03e-219 | 37 | 374 | 4 | 341 | Crystalstructure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3GVE_B Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 2Z1A_A | 2.81e-101 | 665 | 1174 | 26 | 528 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 5EQV_A | 1.08e-95 | 36 | 372 | 1 | 338 | 1.45Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site [Yersinia pestis CO92] |
| 3JYF_A | 1.89e-91 | 40 | 374 | 4 | 339 | ChainA, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578],3JYF_B Chain B, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578] |
| 5H7W_A | 3.26e-72 | 665 | 1174 | 1 | 522 | Crystalstructure of 5'-nucleotidase from venom of Naja atra [Naja atra],5H7W_B Crystal structure of 5'-nucleotidase from venom of Naja atra [Naja atra] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 0.0 | 1 | 1457 | 1 | 1462 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| P44764 | 7.50e-176 | 35 | 651 | 24 | 653 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=cpdB PE=3 SV=1 |
| P08331 | 3.10e-174 | 35 | 651 | 14 | 643 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Escherichia coli (strain K12) OX=83333 GN=cpdB PE=1 SV=2 |
| P26265 | 3.47e-172 | 35 | 622 | 14 | 611 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=cpdB PE=3 SV=2 |
| P53052 | 1.53e-165 | 24 | 651 | 8 | 648 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Yersinia enterocolitica OX=630 GN=cpdB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
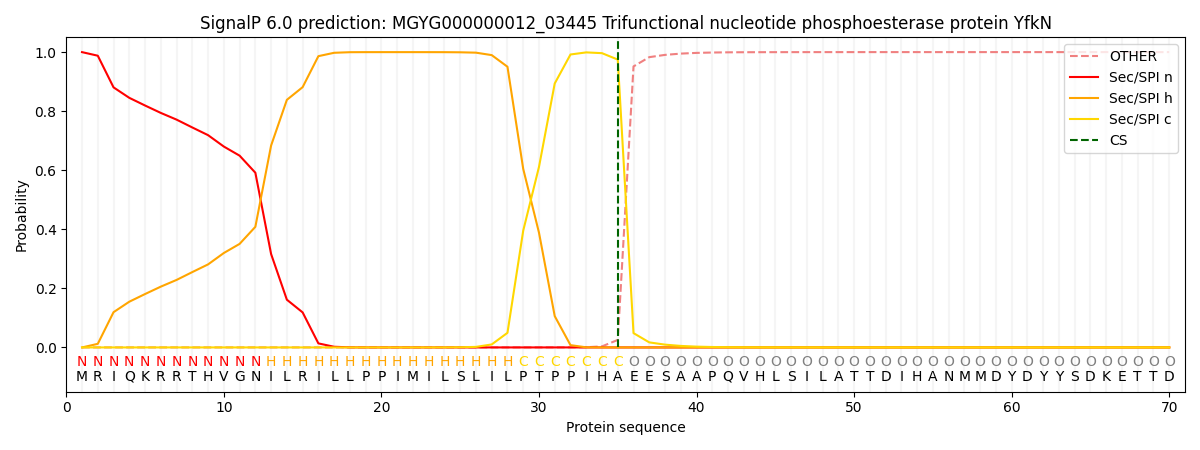
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000573 | 0.998638 | 0.000268 | 0.000178 | 0.000163 | 0.000151 |