You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_00212
Basic Information
help
| Species |
Bacteroides sp902362375
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375
|
| CAZyme ID |
MGYG000000013_00212
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000013 |
6368149 |
Isolate |
United Kingdom |
Europe |
|
| Gene Location |
Start: 286568;
End: 287893
Strand: -
|
No EC number prediction in MGYG000000013_00212.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
74 |
360 |
2e-109 |
0.9884169884169884 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
1.16e-11 |
68 |
320 |
2 |
211 |
Cellulase (glycosyl hydrolase family 5). |
| COG2730
|
BglC |
0.008 |
103 |
282 |
74 |
233 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P06564
|
3.14e-08 |
110 |
313 |
99 |
273 |
Endoglucanase OS=Alkalihalobacillus akibai (strain ATCC 43226 / DSM 21942 / CIP 109018 / JCM 9157 / 1139) OX=1236973 PE=1 SV=1 |
This protein is predicted as LIPO
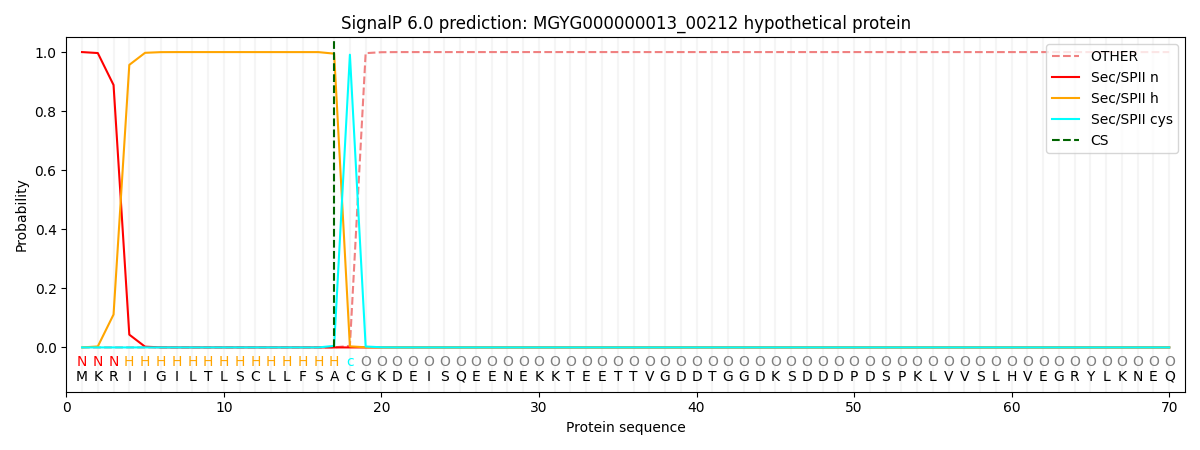
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000000
|
0.000000
|
1.000055
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000000013_00212.

