You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_00235
You are here: Home > Sequence: MGYG000000013_00235
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
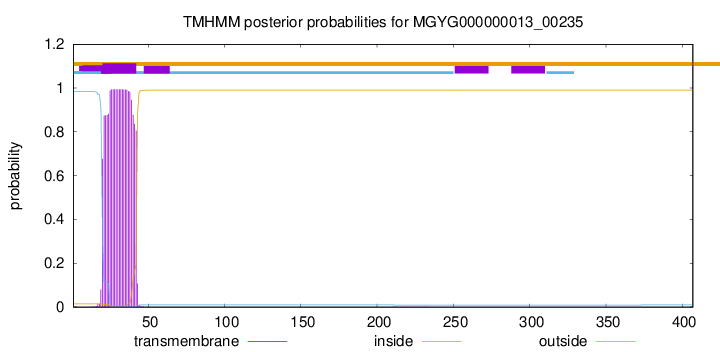
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_00235 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 324712; End: 325935 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 80 | 403 | 5.7e-18 | 0.9739776951672863 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1506 | DAP2 | 3.00e-04 | 241 | 306 | 466 | 533 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0412 | DLH | 5.04e-04 | 141 | 276 | 7 | 140 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam00326 | Peptidase_S9 | 7.08e-04 | 245 | 353 | 61 | 155 | Prolyl oligopeptidase family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW85982.1 | 6.38e-217 | 23 | 403 | 4 | 383 |
| QQA08270.1 | 6.38e-217 | 23 | 403 | 4 | 383 |
| ALJ44193.1 | 6.38e-217 | 23 | 403 | 4 | 383 |
| AAO78713.1 | 6.38e-217 | 23 | 403 | 4 | 383 |
| QDH54042.1 | 1.29e-216 | 23 | 403 | 4 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 2.24e-08 | 86 | 403 | 33 | 366 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
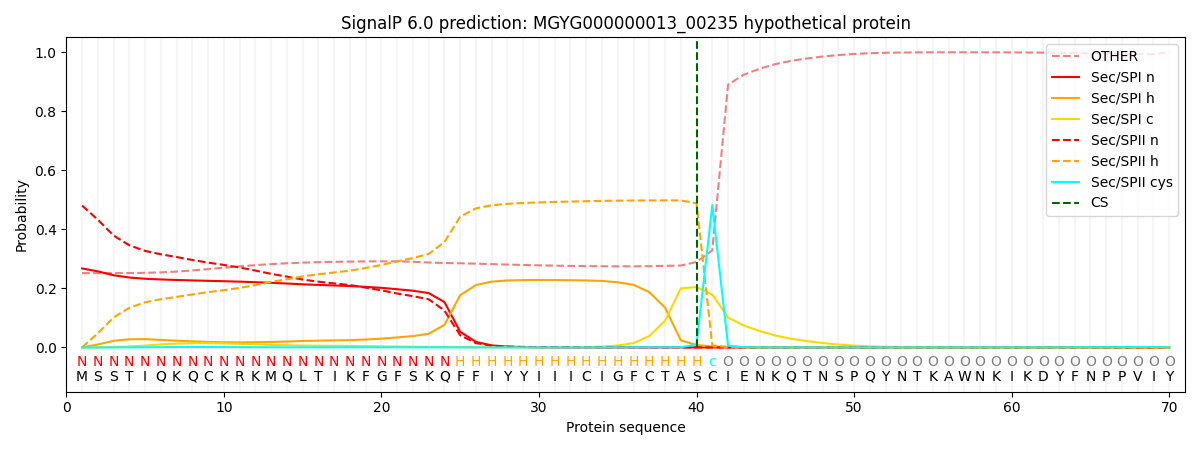
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.257777 | 0.229003 | 0.509635 | 0.000589 | 0.001633 | 0.001356 |