You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_02933
You are here: Home > Sequence: MGYG000000013_02933
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
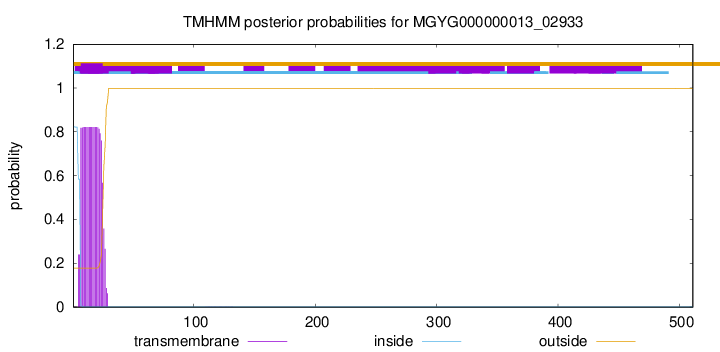
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_02933 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51825; End: 53360 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 93 | 277 | 3.5e-76 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 1.05e-04 | 87 | 277 | 3 | 186 | Amb_all domain. |
| pfam18884 | TSP3_bac | 1.28e-04 | 456 | 477 | 1 | 22 | Bacterial TSP3 repeat. This entry contains a novel bacterial thrombospondin type 3 repeat which differs from the typical consensus by containing a glutamate in place of one of the calcium binding aspartate residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ48964.1 | 0.0 | 3 | 511 | 1 | 509 |
| QRQ55774.1 | 0.0 | 3 | 511 | 1 | 509 |
| QDM11680.1 | 0.0 | 3 | 511 | 1 | 509 |
| QUT78068.1 | 0.0 | 3 | 511 | 1 | 509 |
| QGT73510.1 | 0.0 | 3 | 511 | 1 | 509 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FI2_A | 2.61e-08 | 47 | 189 | 46 | 187 | VexL:A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides [Achromobacter denitrificans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 1.15e-43 | 40 | 502 | 20 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 7.91e-42 | 40 | 502 | 20 | 414 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q4WL88 | 7.53e-40 | 40 | 502 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| B0XMA2 | 7.53e-40 | 40 | 502 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 2.56e-38 | 40 | 502 | 20 | 414 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
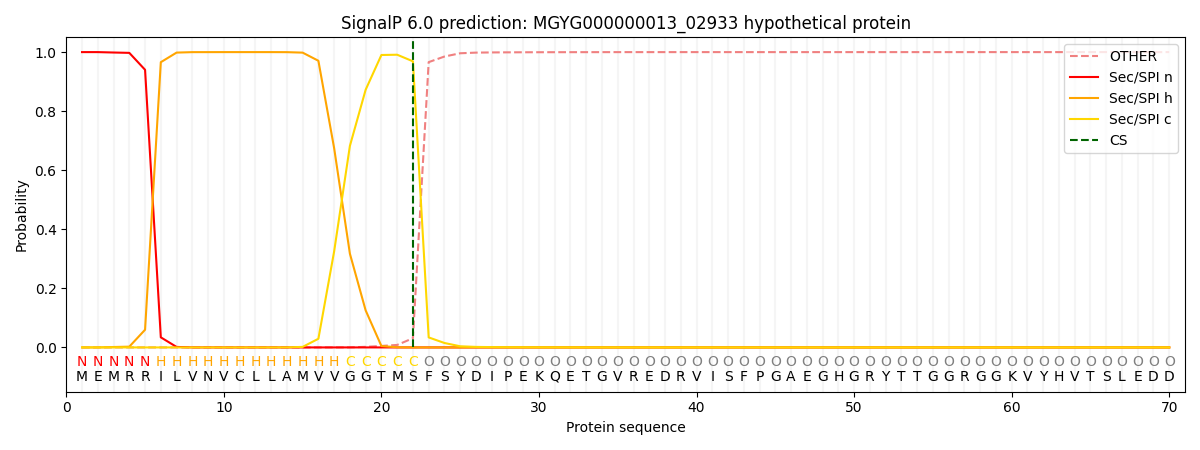
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000315 | 0.999055 | 0.000160 | 0.000154 | 0.000150 | 0.000137 |