You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_04533
You are here: Home > Sequence: MGYG000000013_04533
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_04533 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37454; End: 39406 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 388 | 625 | 3.9e-75 | 0.9781659388646288 |
| CBM51 | 23 | 160 | 6.6e-33 | 0.9925373134328358 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14792 | GH27 | 1.87e-117 | 275 | 558 | 1 | 271 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| PLN02808 | PLN02808 | 1.80e-91 | 271 | 639 | 28 | 376 | alpha-galactosidase |
| PLN02692 | PLN02692 | 1.84e-85 | 251 | 623 | 31 | 384 | alpha-galactosidase |
| PLN02229 | PLN02229 | 8.35e-84 | 236 | 624 | 24 | 394 | alpha-galactosidase |
| pfam16499 | Melibiase_2 | 2.01e-70 | 274 | 558 | 1 | 284 | Alpha galactosidase A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANH83049.1 | 0.0 | 8 | 648 | 9 | 649 |
| BCI63419.1 | 0.0 | 2 | 650 | 8 | 656 |
| AEE50996.1 | 4.15e-201 | 8 | 646 | 13 | 663 |
| AEI50556.1 | 1.84e-199 | 23 | 650 | 44 | 685 |
| QDK80608.1 | 6.25e-198 | 23 | 650 | 27 | 668 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OGZ_A | 2.48e-87 | 187 | 603 | 12 | 429 | Crystalstructure of a putative alpha-galactosidase/melibiase (BF4189) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343],4OGZ_B Crystal structure of a putative alpha-galactosidase/melibiase (BF4189) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343] |
| 4NZJ_A | 2.73e-82 | 187 | 601 | 12 | 427 | Crystalstructure of a putative alpha-galactosidase (BF1418) from Bacteroides fragilis NCTC 9343 at 1.57 A resolution [Bacteroides fragilis NCTC 9343] |
| 1UAS_A | 2.20e-76 | 271 | 620 | 5 | 334 | ChainA, alpha-galactosidase [Oryza sativa] |
| 6F4C_B | 2.41e-75 | 271 | 648 | 5 | 362 | Nicotianabenthamiana alpha-galactosidase [Nicotiana benthamiana] |
| 3A5V_A | 7.83e-59 | 271 | 645 | 5 | 389 | Crystalstructure of alpha-galactosidase I from Mortierella vinacea [Umbelopsis vinacea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14749 | 3.23e-82 | 271 | 648 | 52 | 409 | Alpha-galactosidase OS=Cyamopsis tetragonoloba OX=3832 PE=1 SV=1 |
| Q8RX86 | 5.34e-78 | 271 | 621 | 36 | 366 | Alpha-galactosidase 2 OS=Arabidopsis thaliana OX=3702 GN=AGAL2 PE=1 SV=1 |
| Q9FT97 | 8.50e-77 | 271 | 649 | 50 | 409 | Alpha-galactosidase 1 OS=Arabidopsis thaliana OX=3702 GN=AGAL1 PE=2 SV=1 |
| Q55B10 | 1.59e-76 | 271 | 648 | 24 | 383 | Probable alpha-galactosidase OS=Dictyostelium discoideum OX=44689 GN=melA PE=3 SV=1 |
| Q9FXT4 | 5.91e-75 | 271 | 620 | 60 | 389 | Alpha-galactosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os10g0493600 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
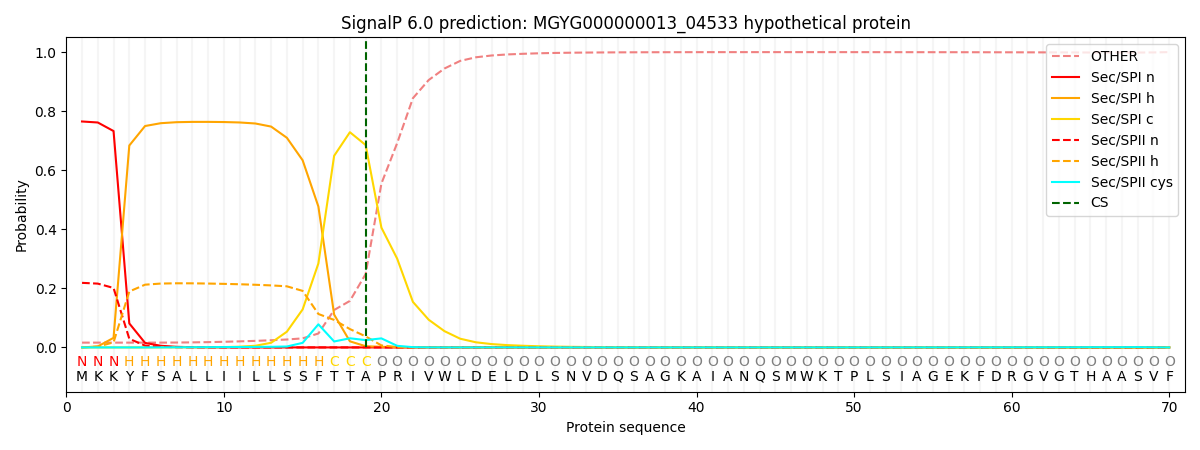
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.019276 | 0.755010 | 0.224692 | 0.000356 | 0.000307 | 0.000317 |
