You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000018_01257
You are here: Home > Sequence: MGYG000000018_01257
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
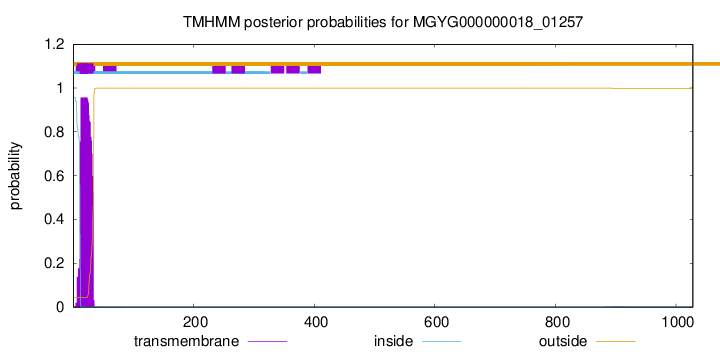
TMHMM annotations
Basic Information help
| Species | Coprococcus eutactus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Coprococcus; Coprococcus eutactus | |||||||||||
| CAZyme ID | MGYG000000018_01257 | |||||||||||
| CAZy Family | CBM61 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9752; End: 12838 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 219 | 596 | 1.7e-126 | 0.9970760233918129 |
| CBM61 | 690 | 828 | 5.7e-26 | 0.9858156028368794 |
| CBM61 | 46 | 181 | 2.3e-18 | 0.9716312056737588 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3867 | GanB | 1.11e-139 | 202 | 608 | 23 | 403 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam07745 | Glyco_hydro_53 | 1.41e-133 | 219 | 596 | 1 | 333 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| pfam07532 | Big_4 | 4.35e-05 | 614 | 671 | 1 | 56 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| TIGR04362 | choice_anch_C | 8.67e-05 | 46 | 181 | 3 | 157 | choice-of-anchor C domain. This family describes an extracellular bacterial domain that occurs on a number of proteins with PEP-CTERM (exosortase recognition site) sequences at the C-terminus, as well some with an apparent alternate anchor sequence. Note that related pfam04862 (DUF642), as of release 26, is double the length of this model because it has two tandem regions homologous to this domain. pfam04862, in turn, belongs to a Pfam clan called the galactose-binding domain-like superfamily. |
| pfam00150 | Cellulase | 0.002 | 232 | 452 | 10 | 214 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK73963.1 | 6.22e-277 | 15 | 835 | 17 | 850 |
| QNM03783.1 | 1.04e-233 | 74 | 840 | 255 | 1091 |
| ADL34775.1 | 2.52e-223 | 54 | 833 | 79 | 877 |
| SQI62631.1 | 2.92e-222 | 4 | 830 | 1 | 808 |
| AYA74678.1 | 3.11e-217 | 4 | 830 | 1 | 810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R8L_A | 3.91e-166 | 195 | 606 | 9 | 398 | Thestructure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1R8L_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1UR0_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR0_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],2CCR_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2CCR_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis] |
| 2GFT_A | 3.07e-165 | 195 | 606 | 9 | 398 | ChainA, Glycosyl Hydrolase Family 53 [Bacillus licheniformis],2GFT_B Chain B, Glycosyl Hydrolase Family 53 [Bacillus licheniformis] |
| 7OSK_A | 4.17e-74 | 213 | 608 | 45 | 403 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1HJS_A | 2.51e-41 | 221 | 536 | 6 | 313 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_A Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus] |
| 1HJQ_A | 2.89e-40 | 220 | 536 | 5 | 313 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65CX5 | 5.16e-165 | 195 | 606 | 34 | 423 | Endo-beta-1,4-galactanase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=ganB PE=1 SV=1 |
| O07013 | 1.43e-159 | 202 | 608 | 37 | 429 | Endo-beta-1,4-galactanase OS=Bacillus subtilis (strain 168) OX=224308 GN=ganB PE=1 SV=1 |
| P48843 | 1.13e-51 | 214 | 526 | 2 | 314 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P48841 | 6.50e-43 | 218 | 525 | 27 | 330 | Arabinogalactan endo-beta-1,4-galactanase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ganB PE=1 SV=1 |
| A1D3T4 | 7.22e-41 | 221 | 536 | 27 | 336 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=galA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
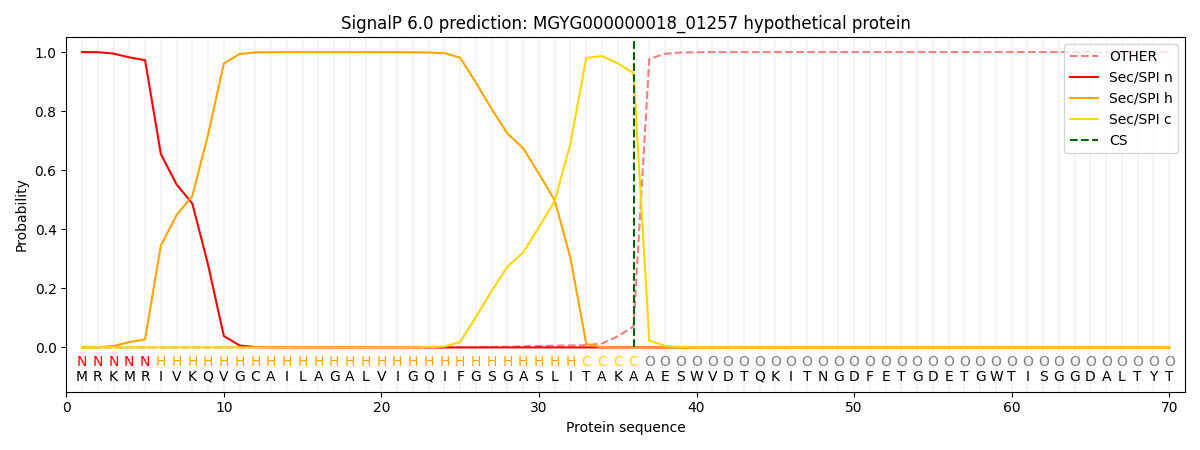
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000519 | 0.998626 | 0.000209 | 0.000246 | 0.000197 | 0.000173 |