You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000019_03315
You are here: Home > Sequence: MGYG000000019_03315
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterobacter cloacae_O | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacter; Enterobacter cloacae_O | |||||||||||
| CAZyme ID | MGYG000000019_03315 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75160; End: 77361 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 24 | 713 | 7.7e-108 | 0.6981382978723404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.71e-42 | 25 | 630 | 14 | 635 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.46e-17 | 65 | 428 | 62 | 418 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.30e-14 | 26 | 427 | 44 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 7.99e-09 | 192 | 306 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK09525 | lacZ | 1.69e-07 | 25 | 427 | 54 | 462 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLA69557.1 | 0.0 | 1 | 733 | 1 | 733 |
| QKZ98086.1 | 0.0 | 1 | 733 | 1 | 733 |
| ASA04277.1 | 0.0 | 4 | 733 | 3 | 732 |
| AZU68037.1 | 0.0 | 4 | 733 | 3 | 732 |
| APR44849.1 | 0.0 | 4 | 733 | 3 | 732 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6U_A | 9.52e-38 | 89 | 654 | 102 | 675 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6BYC_A | 1.64e-33 | 89 | 676 | 91 | 715 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYE_A | 1.64e-33 | 89 | 676 | 91 | 715 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 3.82e-33 | 89 | 676 | 89 | 713 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 8.93e-33 | 89 | 676 | 91 | 715 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5B7W2 | 6.88e-35 | 55 | 627 | 51 | 656 | Beta-mannosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mndB PE=1 SV=2 |
| I2C092 | 1.25e-34 | 69 | 627 | 63 | 660 | Beta-mannosidase B OS=Thermothelomyces thermophilus OX=78579 GN=man9 PE=1 SV=1 |
| Q2TXB7 | 6.73e-34 | 63 | 627 | 52 | 656 | Beta-mannosidase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=mndB PE=3 SV=3 |
| B8NW36 | 8.94e-34 | 63 | 627 | 52 | 656 | Beta-mannosidase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=mndB PE=3 SV=1 |
| Q4WAH4 | 1.19e-33 | 63 | 627 | 52 | 656 | Beta-mannosidase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
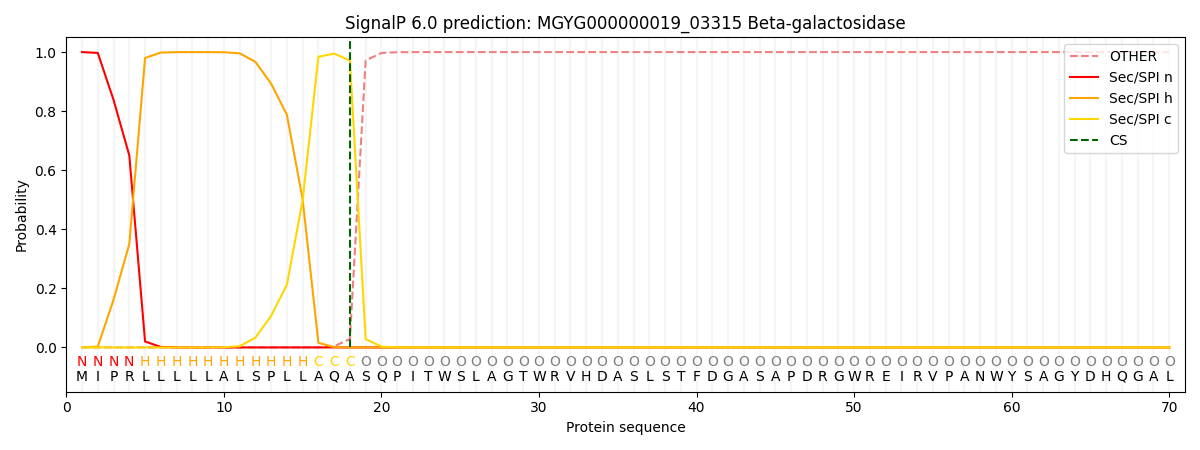
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000447 | 0.998538 | 0.000289 | 0.000255 | 0.000227 | 0.000211 |
