You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000024_00781
You are here: Home > Sequence: MGYG000000024_00781
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraclostridium bifermentans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Paraclostridium; Paraclostridium bifermentans | |||||||||||
| CAZyme ID | MGYG000000024_00781 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 798708; End: 800225 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 334 | 504 | 9e-53 | 0.9491525423728814 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00912 | Transgly | 1.14e-53 | 343 | 504 | 15 | 175 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG0744 | MrcB | 8.34e-47 | 343 | 504 | 78 | 239 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| pfam05569 | Peptidase_M56 | 1.92e-45 | 11 | 294 | 6 | 299 | BlaR1 peptidase M56. Production of beta-Lactamase and penicillin-binding protein 2a (which mediate staphylococcal resistance to beta-lactam antibiotics) is regulated by a signal-transducing integral membrane protein and a transcriptional repressor. The signal transducer is a fusion protein with penicillin-binding and zinc metalloprotease domains. The signal for protein expression is transmitted by site-specific proteolytic cleavage of both the transducer, which auto-activates, and the repressor, which is inactivated, unblocking gene transcription. homologs to this peptidase domain, which corresponds to Merops family M56, are also found in a number of other bacterial genome sequences. |
| cd07341 | M56_BlaR1_MecR1_like | 2.46e-43 | 113 | 298 | 1 | 187 | Peptidase M56-like including those in BlaR1 and MecR1, integral membrane metallopeptidase. This family contains peptidase M56, which includes zinc metalloprotease domain in MecR1 as well as BlaR1. MecR1 is a transmembrane beta-lactam sensor/signal transducer protein that regulates the expression of an altered penicillin-binding protein PBP2a, which resists inactivation by beta-lactam antibiotics, in methicillin-resistant Staphylococcus aureus (MRSA). BlaR1 regulates the inducible expression of a class A beta-lactamase that hydrolytically destroys certain ?-lactam antibiotics in MRSA. Both, MecR1 and BlaR1, are transmembrane proteins that consist of four transmembrane helices, a cytoplasmic zinc protease domain, and the soluble C-terminal extracellular sensor domain, and are highly similar in sequence and function. The signal for protein expression is transmitted by site-specific proteolytic cleavage of both the transducer, which auto-activates, and the repressor, which is inactivated, unblocking gene transcription. All members contain the zinc metalloprotease motif (HEXXH). Homologs of this peptidase domain are also found in a number of other bacterial genome sequences, most of which are as yet uncharacterized. |
| PRK00056 | mtgA | 1.73e-31 | 334 | 503 | 57 | 221 | monofunctional biosynthetic peptidoglycan transglycosylase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFA50087.1 | 2.47e-167 | 1 | 505 | 1 | 506 |
| BAK56295.1 | 2.54e-29 | 322 | 505 | 32 | 213 |
| BAK79624.1 | 2.54e-29 | 322 | 505 | 32 | 213 |
| QJS18329.1 | 2.75e-29 | 332 | 503 | 59 | 229 |
| QVK21516.1 | 4.07e-29 | 343 | 502 | 60 | 218 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NB6_A | 1.09e-26 | 343 | 505 | 22 | 183 | Crystalstructure of Aquifex aeolicus peptidoglycan glycosyltransferase in complex with Methylphosphoryl Neryl Moenomycin [Aquifex aeolicus] |
| 2OQO_A | 1.50e-26 | 343 | 505 | 22 | 183 | Crystalstructure of a peptidoglycan glycosyltransferase from a class A PBP: insight into bacterial cell wall synthesis [Aquifex aeolicus VF5],3D3H_A Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A [Aquifex aeolicus],3NB7_A Crystal structure of Aquifex Aeolicus Peptidoglycan Glycosyltransferase in complex with Decarboxylated Neryl Moenomycin [Aquifex aeolicus] |
| 3HZS_A | 6.36e-20 | 331 | 505 | 10 | 190 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus MW2] |
| 6FTB_A | 6.50e-20 | 331 | 505 | 11 | 191 | Staphylococcusaureus monofunctional glycosyltransferase in complex with moenomycin [Staphylococcus aureus MW2] |
| 5ZZK_A | 3.41e-19 | 331 | 505 | 46 | 226 | ChainA, Monofunctional glycosyltransferase [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38050 | 7.48e-26 | 323 | 502 | 47 | 227 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
| Q51005 | 4.27e-24 | 343 | 505 | 61 | 222 | Biosynthetic peptidoglycan transglycosylase OS=Neisseria gonorrhoeae OX=485 GN=mtgA PE=3 SV=1 |
| Q5F6F6 | 4.27e-24 | 343 | 505 | 61 | 222 | Biosynthetic peptidoglycan transglycosylase OS=Neisseria gonorrhoeae (strain ATCC 700825 / FA 1090) OX=242231 GN=mtgA PE=3 SV=1 |
| B4RJ59 | 5.85e-24 | 343 | 505 | 61 | 222 | Biosynthetic peptidoglycan transglycosylase OS=Neisseria gonorrhoeae (strain NCCP11945) OX=521006 GN=mtgA PE=3 SV=1 |
| O66874 | 1.86e-23 | 343 | 505 | 65 | 226 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000029 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
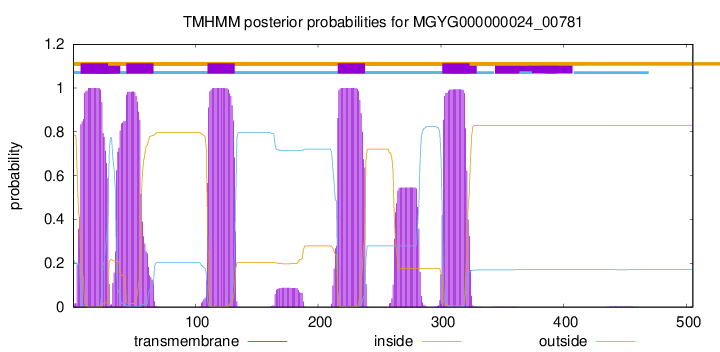
| start | end |
|---|---|
| 7 | 29 |
| 44 | 66 |
| 110 | 132 |
| 216 | 238 |
| 301 | 323 |
