You are browsing environment: HUMAN GUT
MGYG000000024_01321
Basic Information
help
Species
Paraclostridium bifermentans
Lineage
Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Paraclostridium; Paraclostridium bifermentans
CAZyme ID
MGYG000000024_01321
CAZy Family
GH73
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000024
3641600
Isolate
United Kingdom
Europe
Gene Location
Start: 1367949;
End: 1368665
Strand: -
No EC number prediction in MGYG000000024_01321.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam01832
Glucosaminidase
2.78e-07
109
169
2
74
Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan.
more
COG4193
LytD
1.43e-06
31
182
38
189
Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism].
more
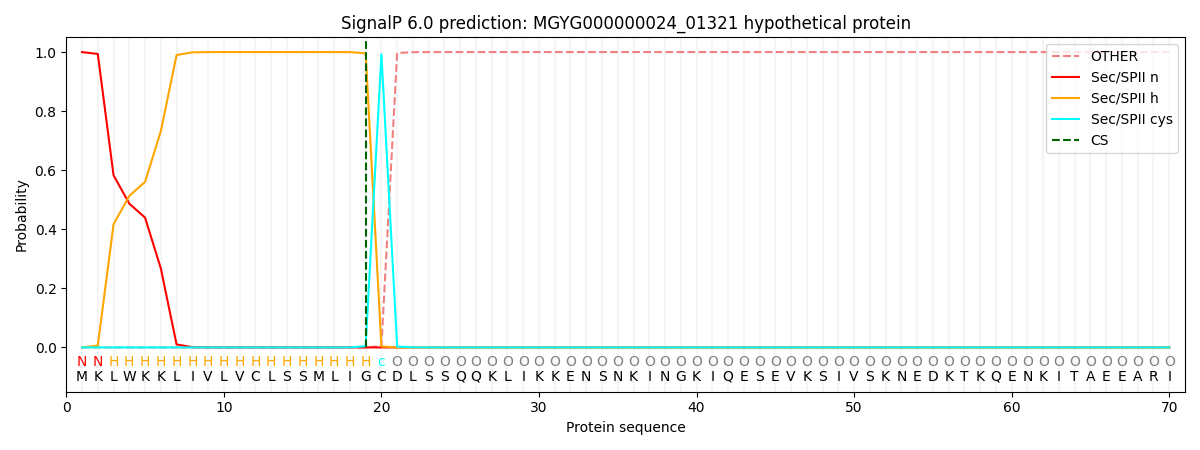
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000000
1.000057
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000000024_01321.